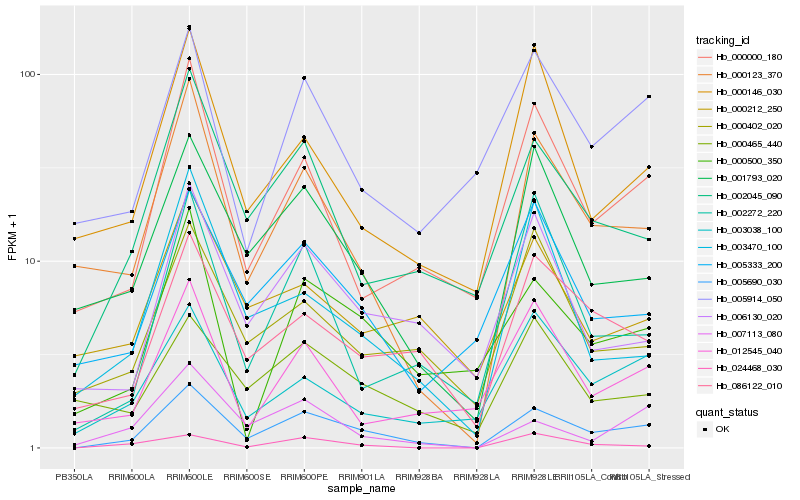
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005690_030 |
0.0 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 2 |
Hb_002045_090 |
0.1817876658 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 3 |
Hb_000123_370 |
0.1819384479 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_086122_010 |
0.1842796644 |
- |
- |
PREDICTED: immunoglobulin-like domain-containing receptor 1 [Jatropha curcas] |
| 5 |
Hb_001793_020 |
0.1864778691 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 6 |
Hb_000000_180 |
0.1910691797 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_006130_020 |
0.1989577553 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000500_350 |
0.2016965901 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 9 |
Hb_005333_200 |
0.2058204504 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_007113_080 |
0.2095672102 |
- |
- |
PREDICTED: uncharacterized protein LOC105639553 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000402_020 |
0.209828223 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_003038_100 |
0.2103716046 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005914_050 |
0.2129101256 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002272_220 |
0.2168520777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000146_030 |
0.216932629 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000465_440 |
0.2169359163 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 17 |
Hb_024468_030 |
0.2169367497 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 18 |
Hb_012545_040 |
0.2200897105 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_003470_100 |
0.2243538035 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000212_250 |
0.2249823882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |