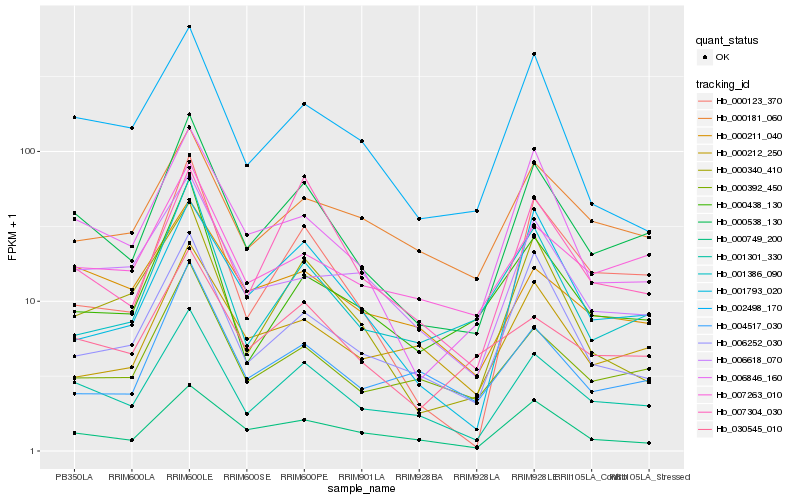
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_130 |
0.0 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 2 |
Hb_001301_330 |
0.0785366704 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 3 |
Hb_006846_160 |
0.1267278849 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 4 |
Hb_000123_370 |
0.1291977237 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000392_450 |
0.1373955414 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000749_200 |
0.1384950973 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_030545_010 |
0.1430533367 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 8 |
Hb_007263_010 |
0.1452207124 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_006252_030 |
0.1469103545 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 10 |
Hb_000438_130 |
0.149030404 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 11 |
Hb_001386_090 |
0.1495662351 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 12 |
Hb_006618_070 |
0.1531330013 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001793_020 |
0.1539152194 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 14 |
Hb_000340_410 |
0.1559242428 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 15 |
Hb_007304_030 |
0.1560639508 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002498_170 |
0.1566542622 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000211_040 |
0.1589065003 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 18 |
Hb_000181_060 |
0.1606093014 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004517_030 |
0.1608392873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000212_250 |
0.1611327967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |