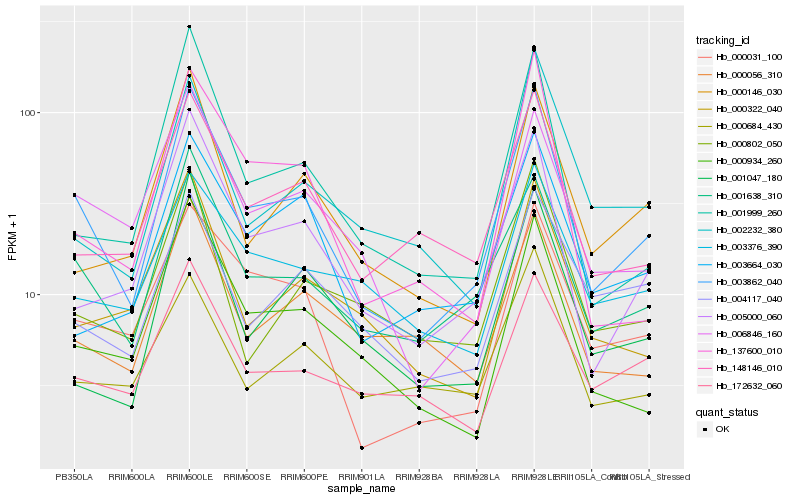
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003862_040 |
0.0 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001638_310 |
0.1082825419 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 3 |
Hb_005000_060 |
0.1298794098 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 4 |
Hb_006846_160 |
0.1304672732 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_000031_100 |
0.1382784296 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 6 |
Hb_148146_010 |
0.1443583079 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 7 |
Hb_000934_260 |
0.146380444 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001999_260 |
0.147536799 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000146_030 |
0.1491622249 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 10 |
Hb_172632_060 |
0.1512532403 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 11 |
Hb_137600_010 |
0.1523681254 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 12 |
Hb_000056_310 |
0.1568458176 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 13 |
Hb_000322_040 |
0.1587733422 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002232_380 |
0.1595108611 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000684_430 |
0.1599125816 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000802_050 |
0.1599317075 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 17 |
Hb_003376_390 |
0.1599922164 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004117_040 |
0.1604698886 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 19 |
Hb_003664_030 |
0.1605525769 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 20 |
Hb_001047_180 |
0.1619070083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |