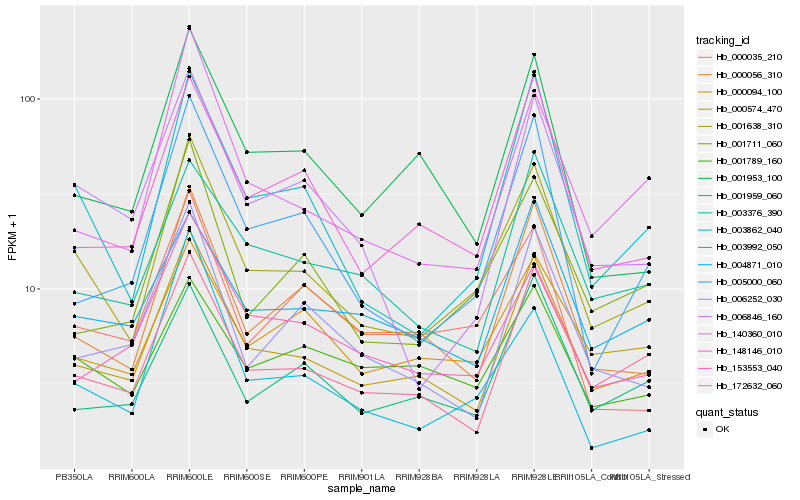
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_310 |
0.0 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 2 |
Hb_003862_040 |
0.1082825419 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000574_470 |
0.1268188682 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001711_060 |
0.1270028657 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 5 |
Hb_000094_100 |
0.1285626446 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 6 |
Hb_172632_060 |
0.1295430036 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000056_310 |
0.1305016555 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 8 |
Hb_148146_010 |
0.1359149964 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 9 |
Hb_140360_010 |
0.1387542146 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_006846_160 |
0.1393043661 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 11 |
Hb_005000_060 |
0.1452679039 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 12 |
Hb_003992_050 |
0.1482313021 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 13 |
Hb_000035_210 |
0.1493698992 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 14 |
Hb_003376_390 |
0.1504657001 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001959_060 |
0.151166339 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_153553_040 |
0.1532742783 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 17 |
Hb_004871_010 |
0.1544075036 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 18 |
Hb_001953_100 |
0.1552523613 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006252_030 |
0.1557040398 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 20 |
Hb_001789_160 |
0.1562232593 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |