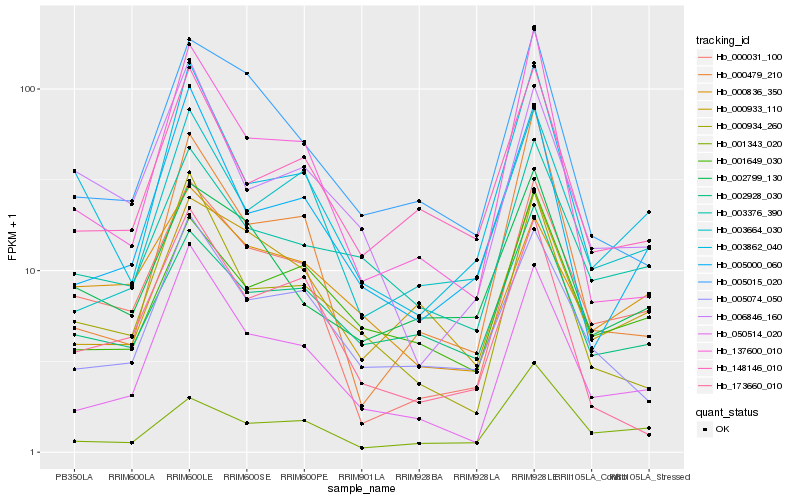
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_100 |
0.0 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 2 |
Hb_003862_040 |
0.1382784296 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_050514_020 |
0.146827389 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 4 |
Hb_006846_160 |
0.1565692705 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_003664_030 |
0.157409822 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 6 |
Hb_002799_130 |
0.1639498612 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 7 |
Hb_001343_020 |
0.1683742891 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000933_110 |
0.1688111984 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 9 |
Hb_000836_350 |
0.1697235651 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003376_390 |
0.1717367324 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 11 |
Hb_148146_010 |
0.1717524225 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 12 |
Hb_137600_010 |
0.171863689 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 13 |
Hb_005074_050 |
0.1718908612 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 14 |
Hb_005000_060 |
0.1720611436 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 15 |
Hb_000934_260 |
0.1728813383 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_005015_020 |
0.1739567516 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000479_210 |
0.1742663626 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 18 |
Hb_002928_030 |
0.1776933377 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 19 |
Hb_001649_030 |
0.1779442767 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_173660_010 |
0.1783989562 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |