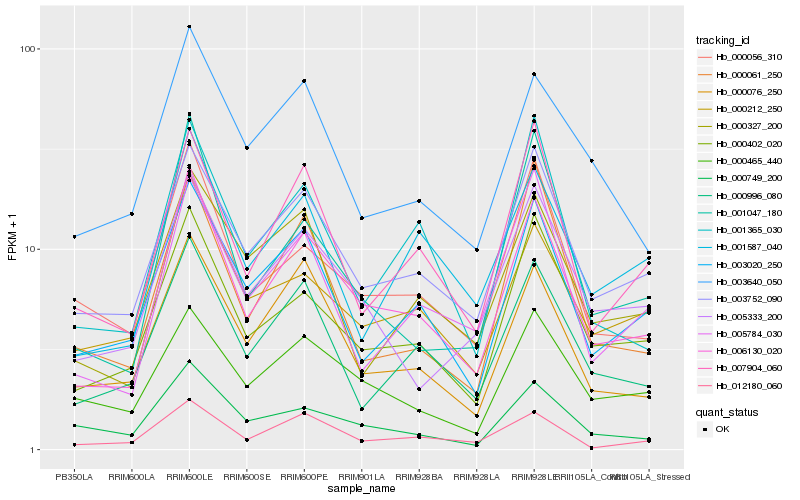
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006130_020 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000402_020 |
0.1070981988 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_003752_090 |
0.1105256657 |
- |
- |
chitinase, putative [Ricinus communis] |
| 4 |
Hb_000076_250 |
0.1219284093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000996_080 |
0.1222905882 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 6 |
Hb_007904_060 |
0.1233172536 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 7 |
Hb_000056_310 |
0.1238351187 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 8 |
Hb_001047_180 |
0.1244731539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003020_250 |
0.1258715808 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 10 |
Hb_012180_060 |
0.1272824341 |
- |
- |
peptidase, putative [Ricinus communis] |
| 11 |
Hb_005333_200 |
0.1302525689 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000212_250 |
0.1322029421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005784_030 |
0.1341986185 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001587_040 |
0.1343258568 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001365_030 |
0.1353124396 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_003640_050 |
0.1376765952 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_000327_200 |
0.1393844253 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 18 |
Hb_000465_440 |
0.1415723877 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 19 |
Hb_000061_250 |
0.1436453355 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 20 |
Hb_000749_200 |
0.1446097467 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |