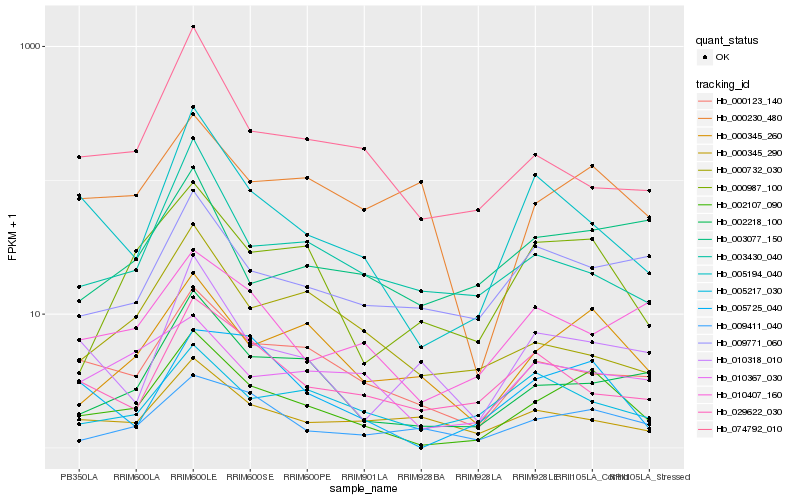
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002107_090 |
0.0 |
transcription factor |
TF Family: bZIP |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 2 |
Hb_000345_260 |
0.1775825724 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_002218_100 |
0.1950286469 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 4 |
Hb_000987_100 |
0.1979188229 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_000123_140 |
0.202530703 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 6 |
Hb_005194_040 |
0.2048635273 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_009411_040 |
0.210483879 |
- |
- |
- |
| 8 |
Hb_005725_040 |
0.2154063558 |
- |
- |
- |
| 9 |
Hb_003430_040 |
0.2333890643 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 10 |
Hb_010367_030 |
0.2339598291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009771_060 |
0.2347709487 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 12 |
Hb_010407_160 |
0.2372102867 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010318_010 |
0.2377698398 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_029622_030 |
0.2403797928 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000345_290 |
0.2406050641 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_074792_010 |
0.2409851439 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 17 |
Hb_000230_480 |
0.2410565204 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase-like [Jatropha curcas] |
| 18 |
Hb_005217_030 |
0.2425447604 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 19 |
Hb_000732_030 |
0.2506383502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003077_150 |
0.2510488231 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |