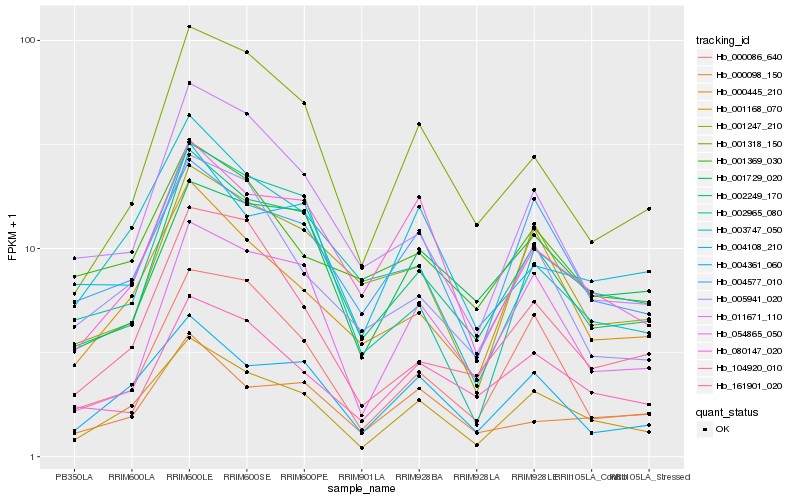
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001168_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 2 |
Hb_004361_060 |
0.1032886153 |
- |
- |
PREDICTED: uncharacterized protein LOC105636705 [Jatropha curcas] |
| 3 |
Hb_004577_010 |
0.1209203501 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 4 |
Hb_000445_210 |
0.1274729313 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 5 |
Hb_002249_170 |
0.1303612047 |
- |
- |
copine, putative [Ricinus communis] |
| 6 |
Hb_080147_020 |
0.1387467757 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 7 |
Hb_000086_640 |
0.1430552143 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_001318_150 |
0.1444087852 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 9 |
Hb_054865_050 |
0.1475601371 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 10 |
Hb_104920_010 |
0.1478596332 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 11 |
Hb_005941_020 |
0.1491684848 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 12 |
Hb_002965_080 |
0.152722582 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 13 |
Hb_001729_020 |
0.1548934306 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_011671_110 |
0.1553547476 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 15 |
Hb_001247_210 |
0.1570552974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001369_030 |
0.1578957149 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_004108_210 |
0.1582563992 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 18 |
Hb_003747_050 |
0.15867909 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 19 |
Hb_000098_150 |
0.1590608493 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 20 |
Hb_161901_020 |
0.1596271441 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |