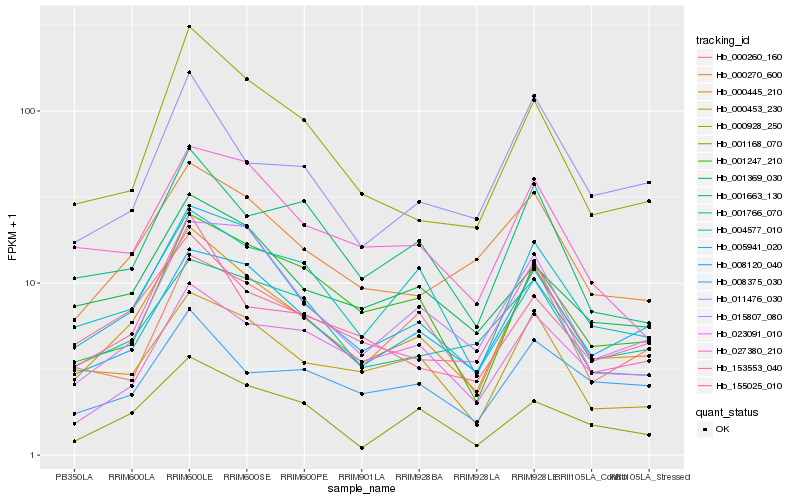
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_210 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 2 |
Hb_008120_040 |
0.1065637224 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 3 |
Hb_153553_040 |
0.1113659952 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 4 |
Hb_004577_010 |
0.1120680091 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 5 |
Hb_000270_600 |
0.1131909443 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 6 |
Hb_155025_010 |
0.121061258 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_015807_080 |
0.1233152809 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 8 |
Hb_000928_250 |
0.123384801 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_001663_130 |
0.1236978168 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 10 |
Hb_001369_030 |
0.1242735144 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_001247_210 |
0.1257953918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000453_230 |
0.1260014084 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_005941_020 |
0.1270471428 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 14 |
Hb_001168_070 |
0.1274729313 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 15 |
Hb_008375_030 |
0.1284351639 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 16 |
Hb_011476_030 |
0.1295625333 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 17 |
Hb_000260_160 |
0.132292789 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_023091_010 |
0.1345180052 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 19 |
Hb_027380_210 |
0.1347042132 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 20 |
Hb_001766_070 |
0.1347047107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |