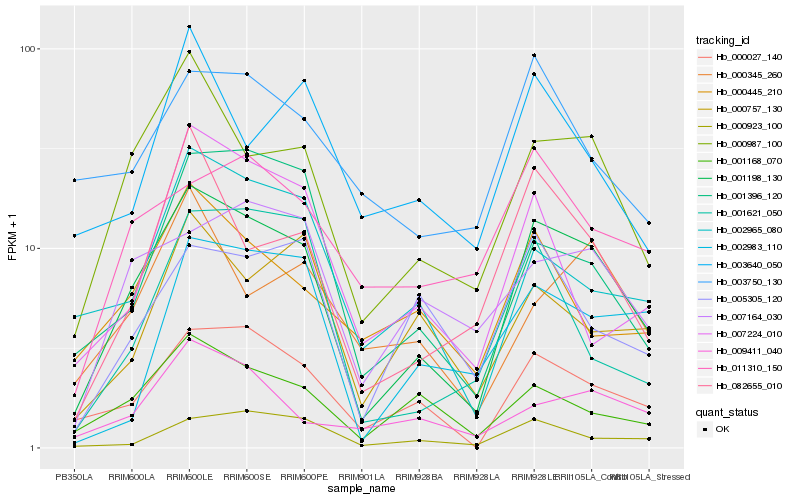
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001198_130 |
0.0 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 2 |
Hb_000987_100 |
0.1468068373 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_000027_140 |
0.1549750519 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 4 |
Hb_001168_070 |
0.1884343179 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 5 |
Hb_082655_010 |
0.1915918135 |
- |
- |
rhodanese-like domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000923_100 |
0.1976486909 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 7 |
Hb_001396_120 |
0.1990312098 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 8 |
Hb_007164_030 |
0.2003462691 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 9 |
Hb_002965_080 |
0.200486782 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 10 |
Hb_005305_120 |
0.2051446439 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 11 |
Hb_000445_210 |
0.2085819179 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 12 |
Hb_011310_150 |
0.2091571617 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 13 |
Hb_000757_130 |
0.2132118527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002983_110 |
0.214957377 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 15 |
Hb_009411_040 |
0.2150281542 |
- |
- |
- |
| 16 |
Hb_007224_010 |
0.2152844917 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 17 |
Hb_003750_130 |
0.2156636602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000345_260 |
0.2172262542 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_001621_050 |
0.21741366 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 20 |
Hb_003640_050 |
0.2216224859 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |