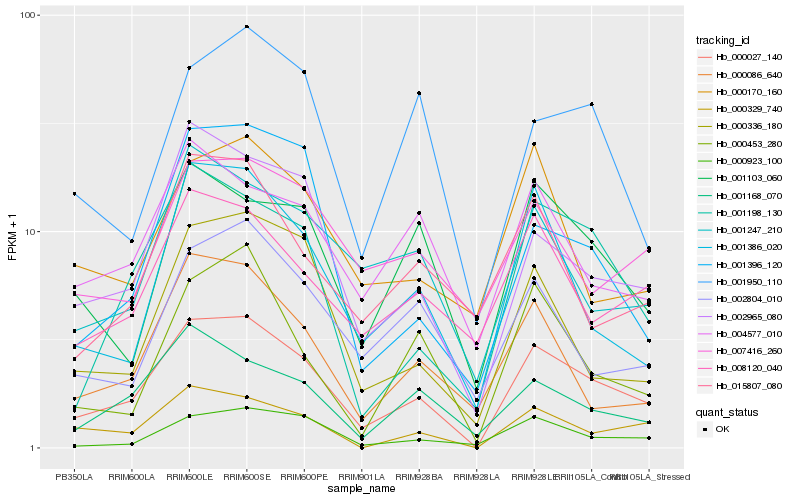
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 2 |
Hb_002965_080 |
0.1394364963 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 3 |
Hb_001198_130 |
0.1549750519 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 4 |
Hb_000329_740 |
0.1556006524 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 5 |
Hb_001386_020 |
0.1572875595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000923_100 |
0.1652706446 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 7 |
Hb_001168_070 |
0.1656118296 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 8 |
Hb_000336_180 |
0.1676598533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001247_210 |
0.1711371017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000086_640 |
0.1711975539 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_001103_060 |
0.1717590706 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 12 |
Hb_008120_040 |
0.1725248435 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 13 |
Hb_000453_280 |
0.1756605929 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 14 |
Hb_001396_120 |
0.1763381281 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 15 |
Hb_004577_010 |
0.1765621185 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 16 |
Hb_002804_010 |
0.1781757222 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 17 |
Hb_007416_260 |
0.1828073346 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 18 |
Hb_000170_160 |
0.183443398 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001950_110 |
0.1845667146 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 20 |
Hb_015807_080 |
0.1849944505 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |