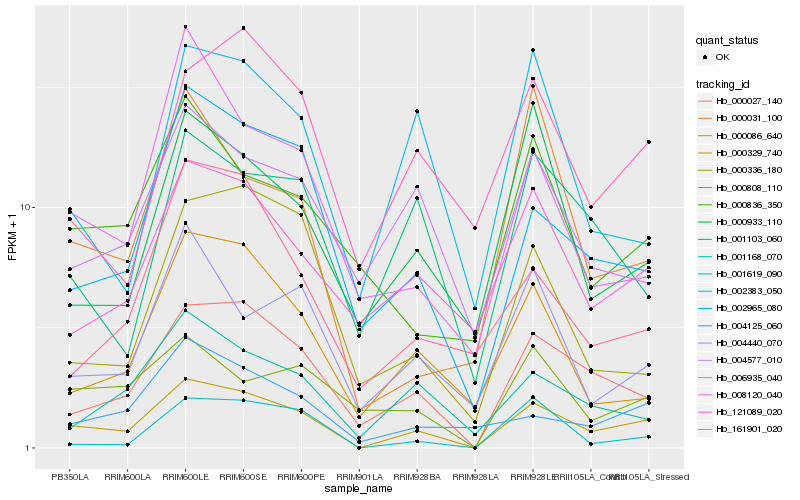
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_740 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 2 |
Hb_000027_140 |
0.1556006524 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 3 |
Hb_002965_080 |
0.1782381568 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 4 |
Hb_001168_070 |
0.2027425916 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 5 |
Hb_000086_640 |
0.2032846657 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_008120_040 |
0.2061619426 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 7 |
Hb_004125_060 |
0.2082489472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004440_070 |
0.2105934616 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 9 |
Hb_006935_040 |
0.2123885491 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 10 |
Hb_000808_110 |
0.2149191576 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000933_110 |
0.215869503 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 12 |
Hb_004577_010 |
0.2168193065 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 13 |
Hb_121089_020 |
0.2178582526 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 14 |
Hb_001619_090 |
0.2183980035 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000836_350 |
0.2187286504 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002383_050 |
0.2188958442 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 17 |
Hb_161901_020 |
0.2198881534 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 18 |
Hb_000336_180 |
0.2206978012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000031_100 |
0.2209092169 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 20 |
Hb_001103_060 |
0.221052157 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |