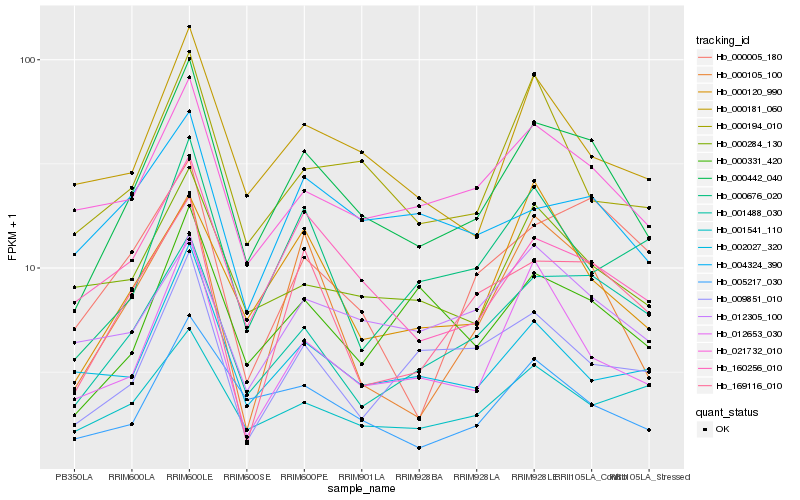
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_040 |
0.0 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_160256_010 |
0.1323228429 |
- |
- |
- |
| 3 |
Hb_005217_030 |
0.1357724239 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 4 |
Hb_021732_010 |
0.1379409874 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 5 |
Hb_000181_060 |
0.138810546 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001488_030 |
0.1394085643 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 7 |
Hb_000331_420 |
0.1432890904 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_169116_010 |
0.147352602 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_012653_030 |
0.1506502438 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_000284_130 |
0.1529691683 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 11 |
Hb_000120_990 |
0.1554114583 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000194_010 |
0.1559353408 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000676_020 |
0.1596463813 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 14 |
Hb_001541_110 |
0.1598822042 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000105_100 |
0.1598989612 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_009851_010 |
0.1627876747 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_012305_100 |
0.1633328786 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002027_320 |
0.1636805806 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 19 |
Hb_004324_390 |
0.1647210505 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 20 |
Hb_000005_180 |
0.1656837075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |