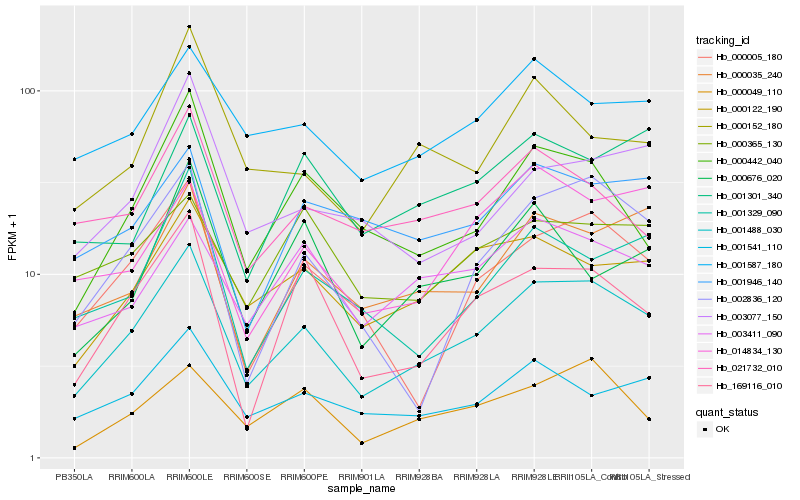
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_030 |
0.0 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 2 |
Hb_000122_190 |
0.1293311973 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000005_180 |
0.1295680639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000035_240 |
0.1315435137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001541_110 |
0.1325934431 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000049_110 |
0.1355625067 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 7 |
Hb_001587_180 |
0.137706353 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 8 |
Hb_169116_010 |
0.1390133195 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_000442_040 |
0.1394085643 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002836_120 |
0.1397942201 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 11 |
Hb_003411_090 |
0.1411255257 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_001329_090 |
0.1423478824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_021732_010 |
0.1516109274 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 14 |
Hb_001946_140 |
0.1520605685 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_014834_130 |
0.1545927255 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_003077_150 |
0.15510525 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 17 |
Hb_000152_180 |
0.1556923076 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 18 |
Hb_000676_020 |
0.1598538288 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 19 |
Hb_001301_340 |
0.1601431314 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 20 |
Hb_000365_130 |
0.160675143 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |