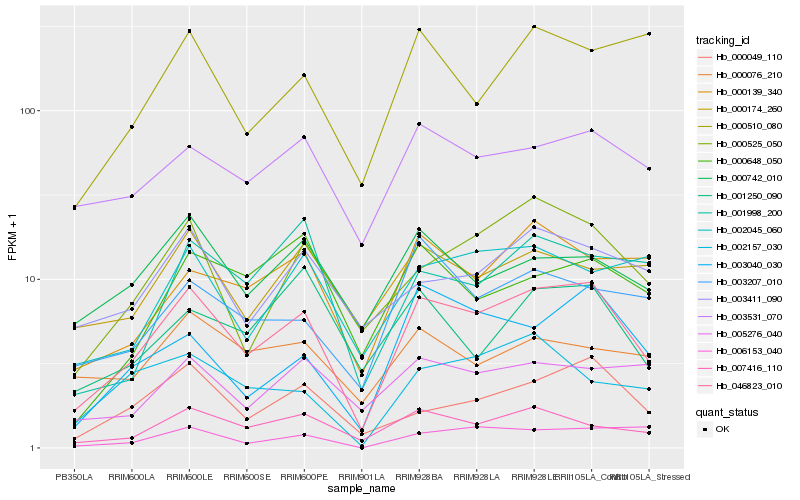
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046823_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 2 |
Hb_007416_110 |
0.1471856983 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000049_110 |
0.1505360738 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 4 |
Hb_000525_050 |
0.1545380747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003531_070 |
0.1627529796 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003040_030 |
0.1684501712 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X1 [Jatropha curcas] |
| 7 |
Hb_000139_340 |
0.1728972753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000742_010 |
0.1730621921 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_003411_090 |
0.1730973292 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_003207_010 |
0.1792468247 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 11 |
Hb_002045_060 |
0.1820893069 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005276_040 |
0.1821955332 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 13 |
Hb_000174_260 |
0.1836817561 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 14 |
Hb_002157_030 |
0.1845643919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001250_090 |
0.187027552 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 16 |
Hb_000648_050 |
0.1879835695 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000510_080 |
0.1883950856 |
- |
- |
hypothetical protein CICLE_v10021453mg [Citrus clementina] |
| 18 |
Hb_006153_040 |
0.1890644869 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 19 |
Hb_001998_200 |
0.1910290119 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 20 |
Hb_000076_210 |
0.1914776436 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |