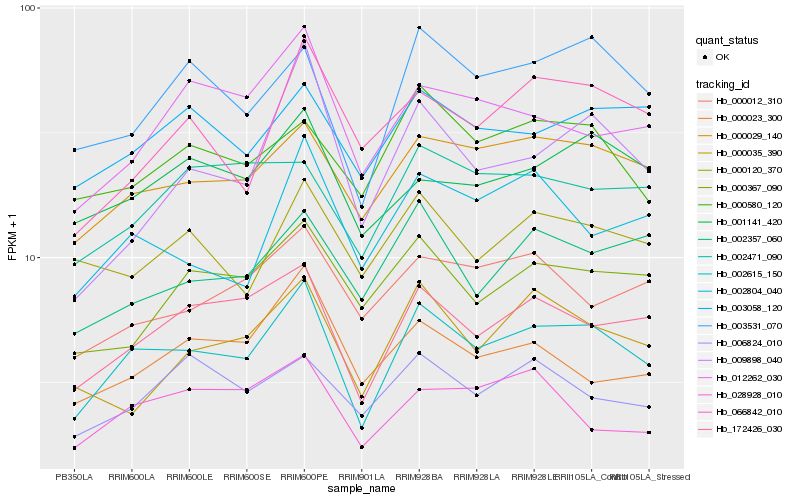
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002615_150 |
0.0 |
- |
- |
PREDICTED: crt homolog 1-like [Jatropha curcas] |
| 2 |
Hb_172426_030 |
0.0920113621 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000029_140 |
0.0948927546 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 4 |
Hb_003531_070 |
0.1010126757 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001141_420 |
0.1169762366 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000580_120 |
0.1177917732 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003058_120 |
0.1177985167 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 8 |
Hb_002804_040 |
0.1207734796 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 9 |
Hb_012262_030 |
0.1211171498 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000367_090 |
0.1213869689 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 11 |
Hb_002357_060 |
0.1216732255 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 12 |
Hb_000035_390 |
0.1219903165 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 13 |
Hb_000012_310 |
0.1234544056 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000120_370 |
0.1244681822 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 15 |
Hb_009898_040 |
0.1248963749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006824_010 |
0.125057194 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 17 |
Hb_000023_300 |
0.1250596718 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 18 |
Hb_002471_090 |
0.1251385785 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 19 |
Hb_066842_010 |
0.1255958579 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_028928_010 |
0.1258133326 |
- |
- |
hypothetical protein CISIN_1g0030132mg, partial [Citrus sinensis] |