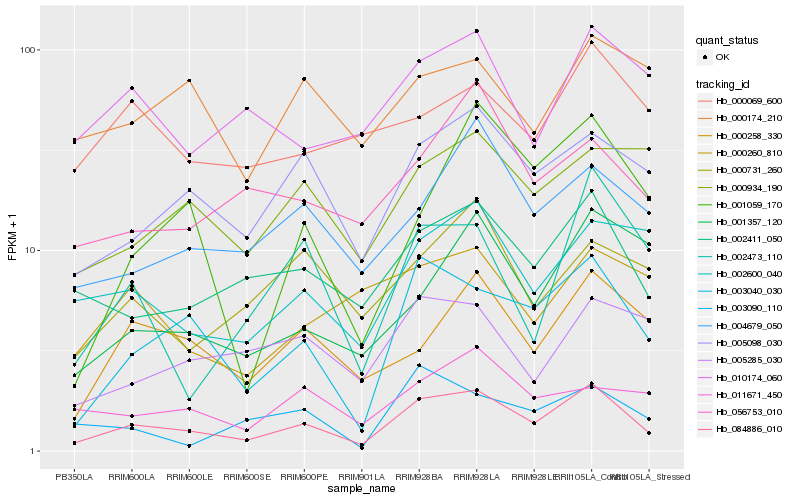
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084886_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003040_030 |
0.1385753552 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X1 [Jatropha curcas] |
| 3 |
Hb_002411_050 |
0.1647968972 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005098_030 |
0.1652001935 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000258_330 |
0.1858658195 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_004679_050 |
0.1876323035 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002600_040 |
0.1902056721 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 8 |
Hb_056753_010 |
0.1994026022 |
- |
- |
PREDICTED: L-galactono-1,4-lactone dehydrogenase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_002473_110 |
0.1995150066 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_000731_260 |
0.2008084927 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 11 |
Hb_010174_060 |
0.2025351827 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_000934_190 |
0.2038046438 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 13 |
Hb_001357_120 |
0.2040780227 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 14 |
Hb_001059_170 |
0.2050367193 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 15 |
Hb_005285_030 |
0.2051034943 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 16 |
Hb_011671_450 |
0.2063790382 |
- |
- |
- |
| 17 |
Hb_003090_110 |
0.21393293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000069_600 |
0.2157045172 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 19 |
Hb_000174_210 |
0.2167854137 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 20 |
Hb_000260_810 |
0.2183472995 |
- |
- |
- |