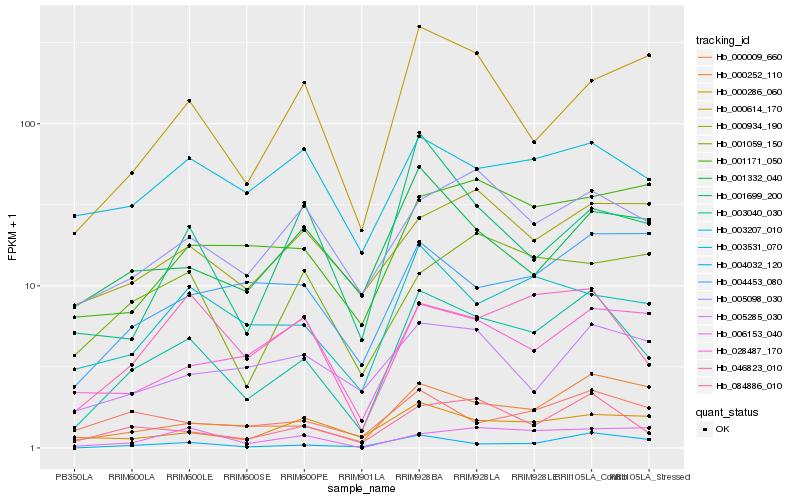
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003040_030 |
0.0 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X1 [Jatropha curcas] |
| 2 |
Hb_084886_010 |
0.1385753552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_046823_010 |
0.1684501712 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 4 |
Hb_000252_110 |
0.1757583473 |
- |
- |
- |
| 5 |
Hb_004032_120 |
0.1817470359 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_003207_010 |
0.1870012957 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 7 |
Hb_005098_030 |
0.1918703772 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000009_660 |
0.2021861167 |
- |
- |
- |
| 9 |
Hb_000286_060 |
0.2052736969 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 10 |
Hb_003531_070 |
0.2094893692 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000614_170 |
0.2105576292 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 12 |
Hb_001332_040 |
0.2114740128 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 13 |
Hb_006153_040 |
0.2129126373 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 14 |
Hb_028487_170 |
0.2139355045 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 15 |
Hb_000934_190 |
0.2158211554 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 16 |
Hb_001171_050 |
0.217478998 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 17 |
Hb_001059_150 |
0.219618711 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 18 |
Hb_004453_080 |
0.2197498318 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 19 |
Hb_005285_030 |
0.2203703533 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 20 |
Hb_001699_200 |
0.2208873726 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |