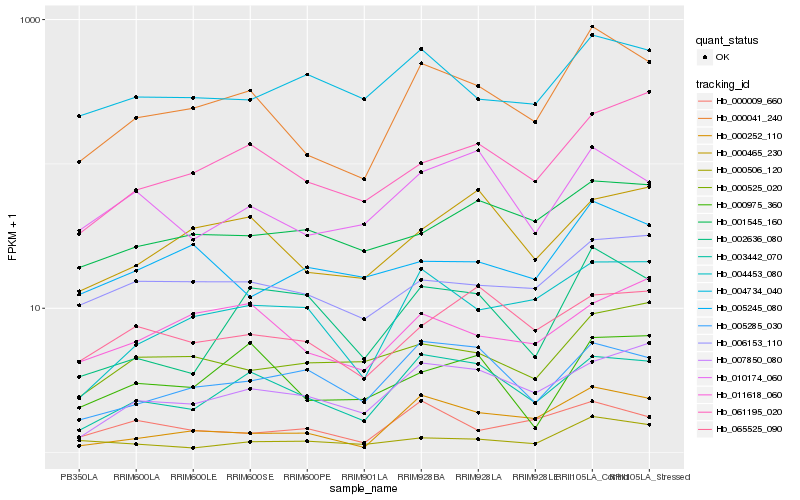
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_240 |
0.0 |
- |
- |
glutaredoxin [Hevea brasiliensis] |
| 2 |
Hb_003442_070 |
0.1359688315 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 3 |
Hb_000252_110 |
0.1440091618 |
- |
- |
- |
| 4 |
Hb_004453_080 |
0.1512318119 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 5 |
Hb_006153_110 |
0.1632799405 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 6 |
Hb_007850_080 |
0.1634193168 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_061195_020 |
0.166044718 |
- |
- |
PREDICTED: uncharacterized protein LOC101990594 [Microtus ochrogaster] |
| 8 |
Hb_000465_230 |
0.1673767532 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 9 |
Hb_010174_060 |
0.1676701244 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 10 |
Hb_000009_660 |
0.1709678199 |
- |
- |
- |
| 11 |
Hb_005285_030 |
0.1722052308 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 12 |
Hb_011618_060 |
0.1755944482 |
- |
- |
PREDICTED: DNA-repair protein XRCC1 [Jatropha curcas] |
| 13 |
Hb_004734_040 |
0.1757331998 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |
| 14 |
Hb_000506_120 |
0.1758879632 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 15 |
Hb_000525_020 |
0.1771413259 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_002636_080 |
0.1775255946 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 17 |
Hb_005245_080 |
0.1776509029 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000975_360 |
0.177919459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_065525_090 |
0.1788440274 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 20 |
Hb_001545_160 |
0.1792273045 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC102615208 isoform X1 [Citrus sinensis] |