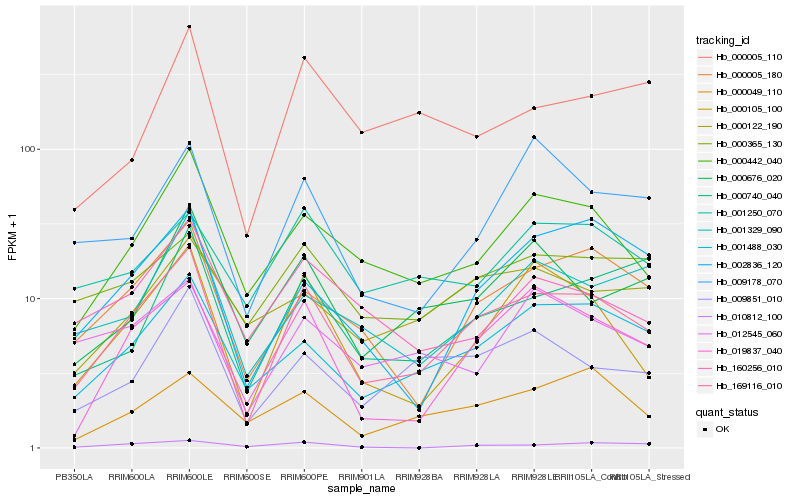
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169116_010 |
0.0 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_001488_030 |
0.1390133195 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 3 |
Hb_019837_040 |
0.1445150122 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 4 |
Hb_000005_180 |
0.1447628293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000105_100 |
0.1466545327 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000442_040 |
0.147352602 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009851_010 |
0.1681024791 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000676_020 |
0.1683939426 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 9 |
Hb_002836_120 |
0.1748064104 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 10 |
Hb_001329_090 |
0.1772750362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000365_130 |
0.1792689776 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 12 |
Hb_000049_110 |
0.1798374275 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 13 |
Hb_000740_040 |
0.1808089093 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 14 |
Hb_009178_070 |
0.1827312429 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 15 |
Hb_160256_010 |
0.1827370205 |
- |
- |
- |
| 16 |
Hb_012545_060 |
0.1831847055 |
- |
- |
PREDICTED: uncharacterized protein LOC103986927 [Musa acuminata subsp. malaccensis] |
| 17 |
Hb_000122_190 |
0.1867725068 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001250_070 |
0.1896740138 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 19 |
Hb_000005_110 |
0.1925665226 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 20 |
Hb_010812_100 |
0.1930015003 |
- |
- |
phosphate transporter [Manihot esculenta] |