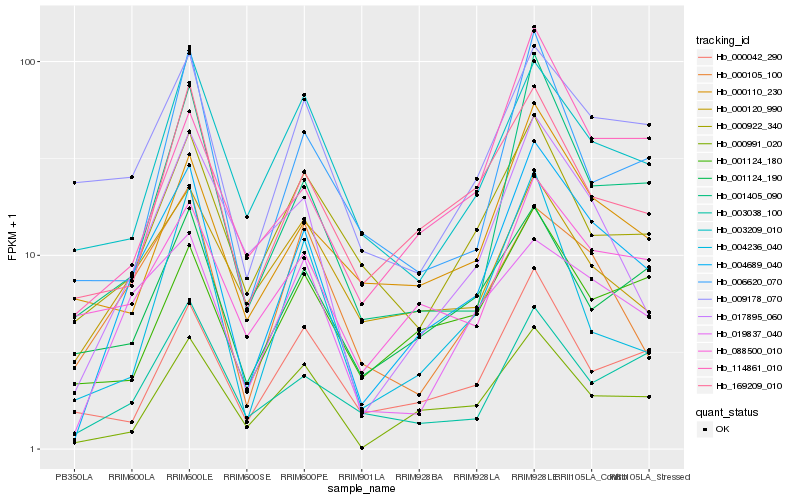
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004689_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 2 |
Hb_017895_060 |
0.1548912237 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 14, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000991_020 |
0.1579592882 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_001405_090 |
0.1605788132 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_019837_040 |
0.1607272526 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 6 |
Hb_169209_010 |
0.1773483705 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009178_070 |
0.1803132557 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000110_230 |
0.1821208847 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_088500_010 |
0.1859282174 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 10 |
Hb_003038_100 |
0.1871960769 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000042_290 |
0.1879089526 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 12 |
Hb_006620_070 |
0.1881616832 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 13 |
Hb_000105_100 |
0.1893593717 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_114861_010 |
0.1893724013 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 15 |
Hb_000922_340 |
0.1897615087 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 16 |
Hb_003209_010 |
0.1917194683 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 17 |
Hb_004236_040 |
0.1920133457 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 18 |
Hb_001124_180 |
0.1932125269 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 19 |
Hb_000120_990 |
0.1949226831 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001124_190 |
0.195070297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |