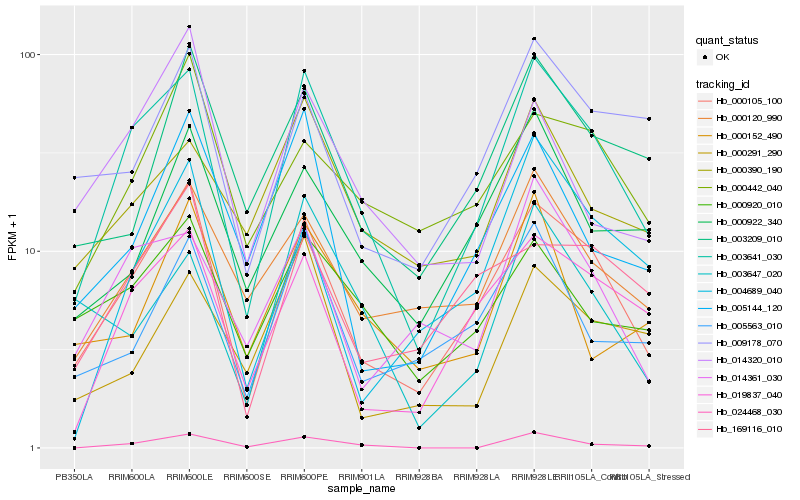
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003641_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 2 |
Hb_000105_100 |
0.1081150117 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_019837_040 |
0.1644770247 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 4 |
Hb_000120_990 |
0.1784228197 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_014361_030 |
0.1822120186 |
- |
- |
hypothetical protein POPTR_0005s18860g [Populus trichocarpa] |
| 6 |
Hb_000390_190 |
0.1842515211 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_024468_030 |
0.185317866 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 8 |
Hb_000920_010 |
0.1927220744 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 9 |
Hb_005563_010 |
0.1936997219 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 10 |
Hb_005144_120 |
0.198163377 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 11 |
Hb_000922_340 |
0.1987508571 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 12 |
Hb_003209_010 |
0.2007858366 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 13 |
Hb_009178_070 |
0.2053726016 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003647_020 |
0.2095591759 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 15 |
Hb_169116_010 |
0.2122601524 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_000291_290 |
0.2131866024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004689_040 |
0.2139035838 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 18 |
Hb_000152_490 |
0.2145849307 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 19 |
Hb_000442_040 |
0.2160355196 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_014320_010 |
0.2179444392 |
- |
- |
PREDICTED: uncharacterized protein LOC105629932 [Jatropha curcas] |