| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
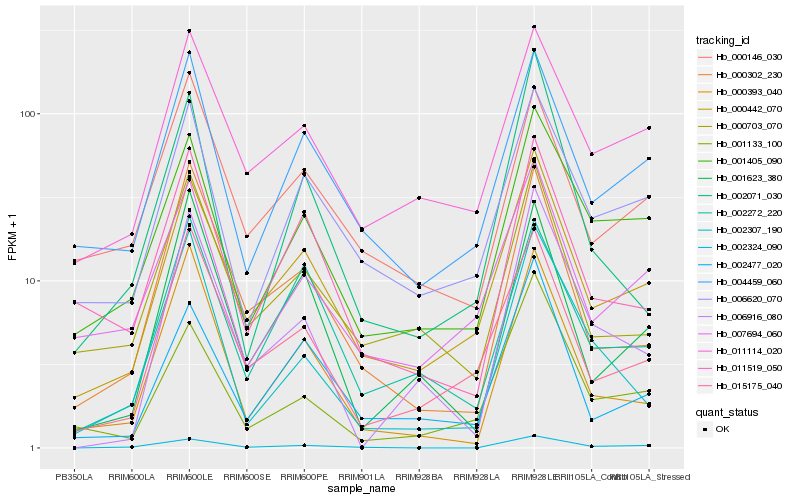
Hb_002324_090 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 2 |
Hb_001405_090 |
0.1612720769 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000442_070 |
0.1682065916 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 4 |
Hb_000302_230 |
0.181758886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004459_060 |
0.1836883203 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000393_040 |
0.1850640892 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 7 |
Hb_006620_070 |
0.1883788591 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 8 |
Hb_002307_190 |
0.1972507791 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 9 |
Hb_011519_050 |
0.1983844254 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 10 |
Hb_006916_080 |
0.1999924411 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 11 |
Hb_011114_020 |
0.2010180831 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_007694_060 |
0.2038279233 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000146_030 |
0.2039551323 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001623_380 |
0.2050627309 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002272_220 |
0.2067881201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002477_020 |
0.2073998035 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 17 |
Hb_001133_100 |
0.2098770006 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 18 |
Hb_015175_040 |
0.2113733943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000703_070 |
0.2139370066 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002071_030 |
0.21439243 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |