| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
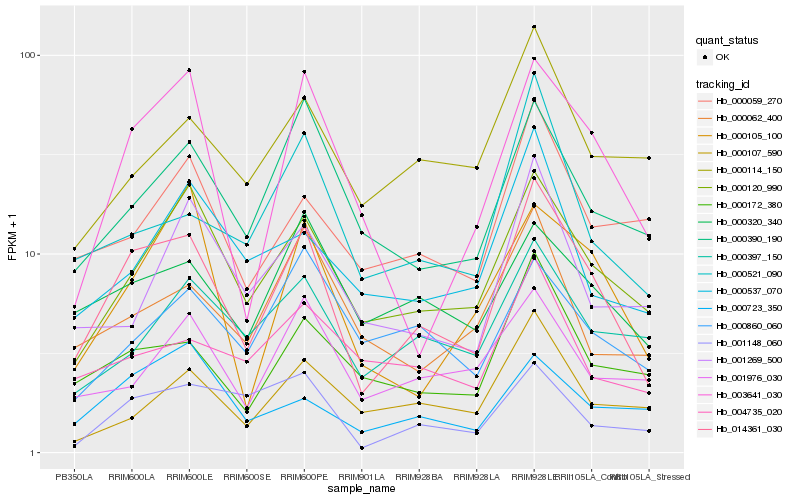
Hb_014361_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s18860g [Populus trichocarpa] |
| 2 |
Hb_000120_990 |
0.1534533171 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000172_380 |
0.1760331971 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 4 |
Hb_000390_190 |
0.180152254 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003641_030 |
0.1822120186 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 6 |
Hb_000320_340 |
0.188415986 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 7 |
Hb_000521_090 |
0.1886220556 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001148_060 |
0.1918177382 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 9 |
Hb_000397_150 |
0.1948050816 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 10 |
Hb_000059_270 |
0.1981952169 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 11 |
Hb_000062_400 |
0.1997233906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000114_150 |
0.2001739612 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000105_100 |
0.2010797456 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001976_030 |
0.201472889 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_004735_020 |
0.2014985507 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 16 |
Hb_000537_070 |
0.2019412409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 17 |
Hb_000107_590 |
0.2034960183 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 18 |
Hb_000860_060 |
0.2060361523 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 19 |
Hb_001269_500 |
0.206473454 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000723_350 |
0.2069067662 |
- |
- |
protein binding protein, putative [Ricinus communis] |