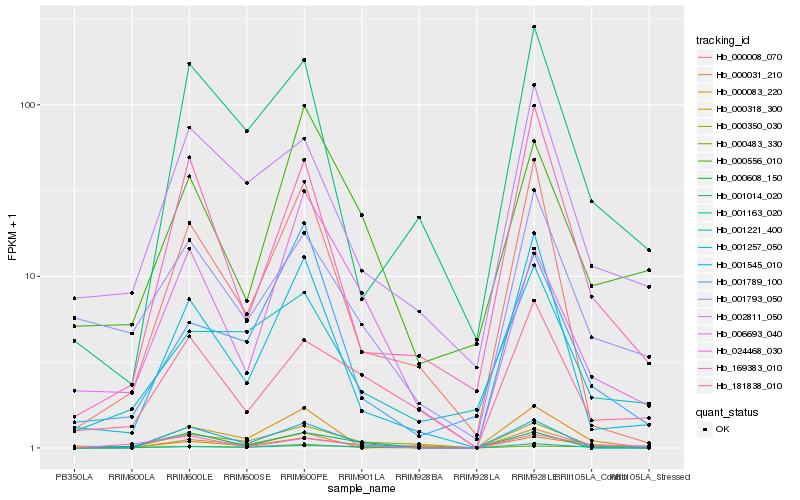
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_300 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105634339 [Jatropha curcas] |
| 2 |
Hb_000483_330 |
0.1711441376 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 3 |
Hb_000350_030 |
0.22597648 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 4 |
Hb_024468_030 |
0.2295790942 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 5 |
Hb_000608_150 |
0.230752848 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001545_010 |
0.2339540578 |
- |
- |
PREDICTED: serine/threonine-protein kinase TNNI3K-like [Cucumis sativus] |
| 7 |
Hb_000008_070 |
0.2400441625 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_001789_100 |
0.2407645068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006693_040 |
0.2421233644 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 10 |
Hb_000083_220 |
0.2434385991 |
- |
- |
- |
| 11 |
Hb_002811_050 |
0.2446726505 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 12 |
Hb_169383_010 |
0.2473440672 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001257_050 |
0.24758159 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 14 |
Hb_181838_010 |
0.2478038972 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 15 |
Hb_001221_400 |
0.2480189405 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 16 |
Hb_000556_010 |
0.2480385439 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001793_050 |
0.250933156 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 18 |
Hb_001163_020 |
0.2546326139 |
desease resistance |
Gene Name: ABC_trans_N |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 19 |
Hb_001014_020 |
0.2558346632 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 20 |
Hb_000031_210 |
0.2567177365 |
- |
- |
cytochrome P450, putative [Ricinus communis] |