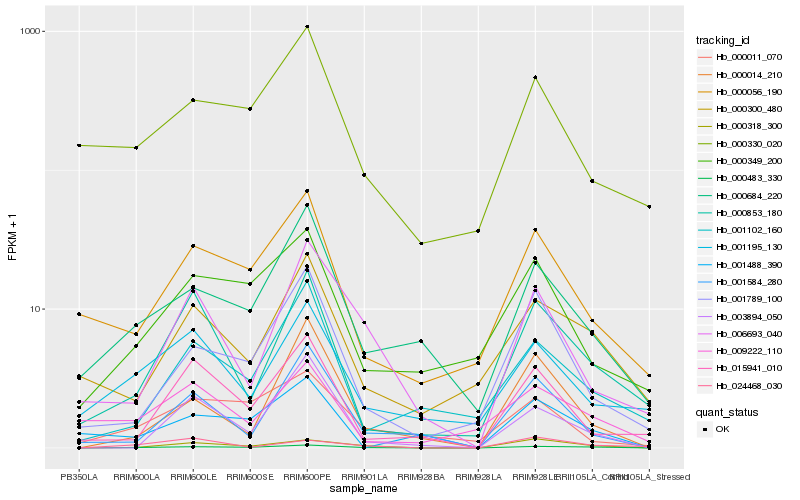
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_330 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 2 |
Hb_000318_300 |
0.1711441376 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105634339 [Jatropha curcas] |
| 3 |
Hb_000684_220 |
0.2343310809 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000300_480 |
0.2453788316 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 5 |
Hb_000853_180 |
0.2488231836 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 6 |
Hb_001789_100 |
0.2490448443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000056_190 |
0.2496284705 |
- |
- |
hypothetical protein [Ricinus communis] |
| 8 |
Hb_000349_200 |
0.2587578596 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g12500 [Jatropha curcas] |
| 9 |
Hb_000011_070 |
0.2626954519 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 10 |
Hb_001102_160 |
0.2642034172 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 11 |
Hb_006693_040 |
0.2670654817 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 12 |
Hb_000330_020 |
0.2671765836 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 13 |
Hb_003894_050 |
0.2680555038 |
- |
- |
hypothetical protein JCGZ_14524 [Jatropha curcas] |
| 14 |
Hb_000014_210 |
0.2682461961 |
- |
- |
amine oxidase, partial [Aphanizomenon flos-aquae] |
| 15 |
Hb_001195_130 |
0.2702264976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001584_280 |
0.2704552466 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 17 |
Hb_009222_110 |
0.2761102505 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 18 |
Hb_024468_030 |
0.2767798316 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 19 |
Hb_015941_010 |
0.2774288788 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_001488_390 |
0.2784674818 |
- |
- |
SP1L, putative [Ricinus communis] |