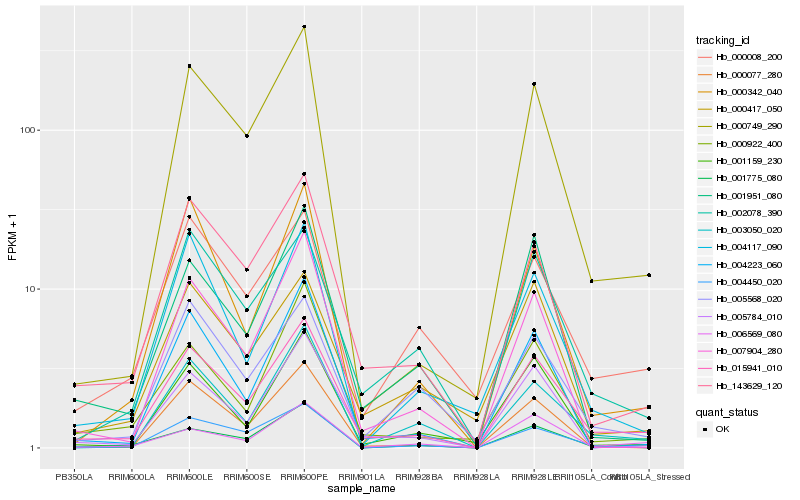
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015941_010 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_143629_120 |
0.118987932 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_001775_080 |
0.1255124348 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 4 |
Hb_000749_290 |
0.1285655083 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 5 |
Hb_002078_390 |
0.1339892696 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 6 |
Hb_005784_010 |
0.1371724961 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 7 |
Hb_001951_080 |
0.1442790402 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000922_400 |
0.1467237191 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 9 |
Hb_006569_080 |
0.1473331988 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 10 |
Hb_004223_060 |
0.147563868 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 11 |
Hb_003050_020 |
0.1480416911 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 12 |
Hb_001159_230 |
0.1481755483 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_000417_050 |
0.1499928034 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 14 |
Hb_004450_020 |
0.1502554813 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000342_040 |
0.1502623137 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 16 |
Hb_004117_090 |
0.1504156204 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 17 |
Hb_005568_020 |
0.1513596321 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000008_200 |
0.1515607495 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 19 |
Hb_000077_280 |
0.1525662905 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_007904_280 |
0.1528261595 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |