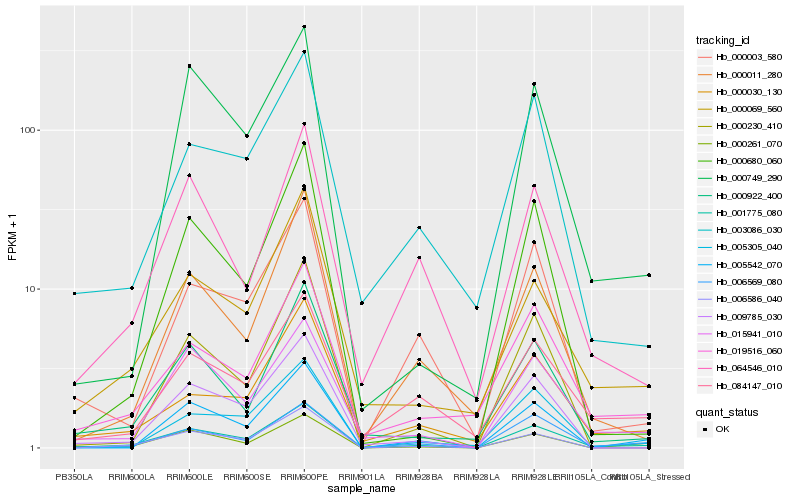
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_080 |
0.0 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 2 |
Hb_006569_080 |
0.1205937638 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 3 |
Hb_015941_010 |
0.1255124348 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_019516_060 |
0.1379206902 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 5 |
Hb_000069_560 |
0.1379753707 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 6 |
Hb_000922_400 |
0.142353161 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 7 |
Hb_005305_040 |
0.1449807087 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000030_130 |
0.1451103363 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_084147_010 |
0.1454138421 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000011_280 |
0.1454629831 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 11 |
Hb_005542_070 |
0.1456141357 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 12 |
Hb_009785_030 |
0.1473851891 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 13 |
Hb_000230_410 |
0.1474638454 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |
| 14 |
Hb_000749_290 |
0.1486553412 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 15 |
Hb_000003_580 |
0.1494641362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006586_040 |
0.1501288873 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |
| 17 |
Hb_000680_060 |
0.1507834598 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 18 |
Hb_064546_010 |
0.1525283403 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 19 |
Hb_000261_070 |
0.1532382469 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 20 |
Hb_003086_030 |
0.1536412761 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |