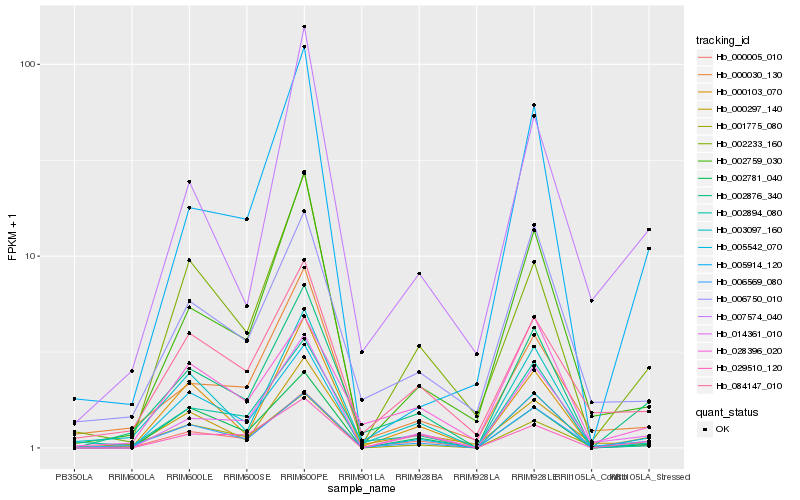
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_340 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s11770g [Populus trichocarpa] |
| 2 |
Hb_006569_080 |
0.1175822895 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 3 |
Hb_000103_070 |
0.1635718754 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 4 |
Hb_003097_160 |
0.1711182423 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 5 |
Hb_028396_020 |
0.1721908902 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 6 |
Hb_084147_010 |
0.1734417635 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_005542_070 |
0.173624108 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 8 |
Hb_002233_160 |
0.1737940906 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 9 |
Hb_000005_010 |
0.1744647062 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 10 |
Hb_014361_010 |
0.1748163828 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_001775_080 |
0.1763004891 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 12 |
Hb_002894_080 |
0.1766059533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_029510_120 |
0.1776665742 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 14 |
Hb_000030_130 |
0.1818753799 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002781_040 |
0.1822551414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000297_140 |
0.1823647484 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 17 |
Hb_006750_010 |
0.1825844152 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 18 |
Hb_002759_030 |
0.1836921535 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 19 |
Hb_005914_120 |
0.185393836 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 20 |
Hb_007574_040 |
0.1861349284 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |