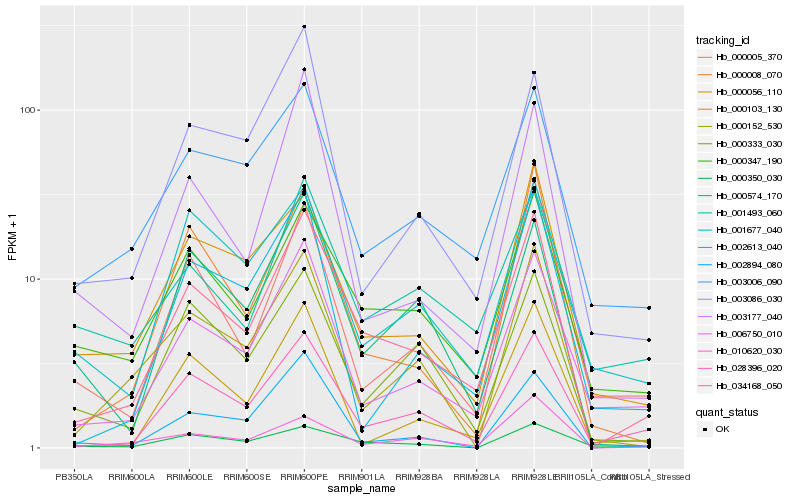
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034168_050 |
0.0 |
- |
- |
Photosystem I assembly protein Ycf3 [Medicago truncatula] |
| 2 |
Hb_028396_020 |
0.1224435058 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 3 |
Hb_006750_010 |
0.1350636487 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_000347_190 |
0.1422578468 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 5 |
Hb_003006_090 |
0.1564321289 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 6 |
Hb_000008_070 |
0.1574856791 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_000056_110 |
0.1613820457 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 8 |
Hb_010620_030 |
0.1676687593 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 9 |
Hb_000574_170 |
0.1678569496 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 10 |
Hb_002894_080 |
0.1678971238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000333_030 |
0.1681541518 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000005_370 |
0.1684935925 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 13 |
Hb_000152_530 |
0.1709999032 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 14 |
Hb_000350_030 |
0.1724989375 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 15 |
Hb_003177_040 |
0.1727410608 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 16 |
Hb_000103_130 |
0.1753024255 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 17 |
Hb_002613_040 |
0.1760985165 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 18 |
Hb_001677_040 |
0.1767833437 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001493_060 |
0.1768568446 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 20 |
Hb_003086_030 |
0.178214703 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |