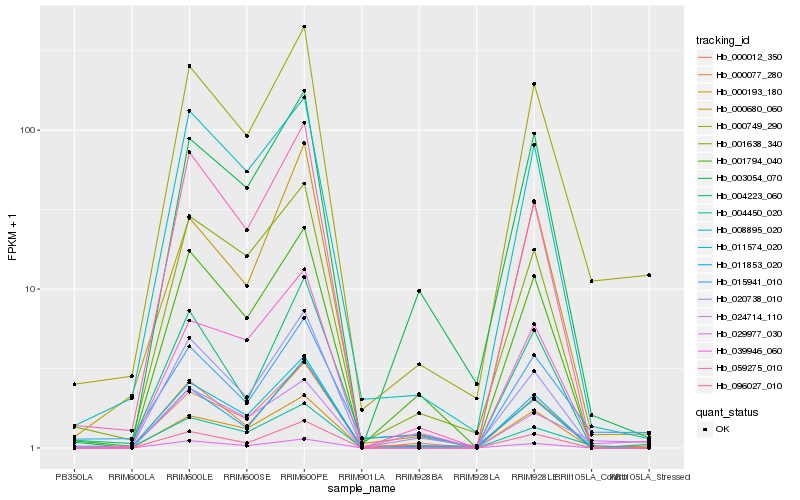
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 2 |
Hb_000077_280 |
0.1096847206 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_039946_060 |
0.1124586517 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 4 |
Hb_011574_020 |
0.1171694599 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 5 |
Hb_024714_110 |
0.1211674351 |
- |
- |
PREDICTED: translocator protein homolog [Jatropha curcas] |
| 6 |
Hb_008895_020 |
0.1217756312 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 7 |
Hb_000193_180 |
0.1222153089 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 8 |
Hb_000012_350 |
0.1238433642 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 9 |
Hb_096027_010 |
0.1259357529 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_011853_020 |
0.1259857563 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 11 |
Hb_059275_010 |
0.126619034 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 12 |
Hb_004450_020 |
0.1272036686 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 13 |
Hb_020738_010 |
0.1278859785 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 14 |
Hb_015941_010 |
0.1285655083 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_001638_340 |
0.1321768468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_029977_030 |
0.1335211572 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000680_060 |
0.1342889445 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 18 |
Hb_003054_070 |
0.1351364759 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001794_040 |
0.137584874 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004223_060 |
0.1383043907 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |