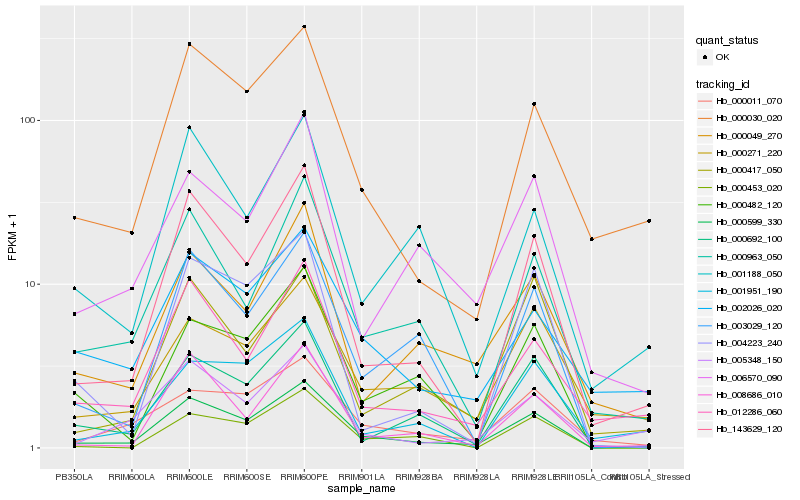
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_330 |
0.0 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 2 |
Hb_143629_120 |
0.1133964532 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_000030_020 |
0.1360897777 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 4 |
Hb_000963_050 |
0.1404142141 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 5 |
Hb_012286_060 |
0.1427187185 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_003029_120 |
0.144650597 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 7 |
Hb_000011_070 |
0.1556167307 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 8 |
Hb_000692_100 |
0.156267389 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000271_220 |
0.1565174274 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_000049_270 |
0.1592877953 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000482_120 |
0.1595525026 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 12 |
Hb_008686_010 |
0.1602203127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002026_020 |
0.1606911413 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 14 |
Hb_005348_150 |
0.1623082924 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 15 |
Hb_006570_090 |
0.1643991852 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 16 |
Hb_000417_050 |
0.1670295743 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 17 |
Hb_001188_050 |
0.167484178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000453_020 |
0.1688091403 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001951_190 |
0.1690614472 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 20 |
Hb_004223_240 |
0.1727747588 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |