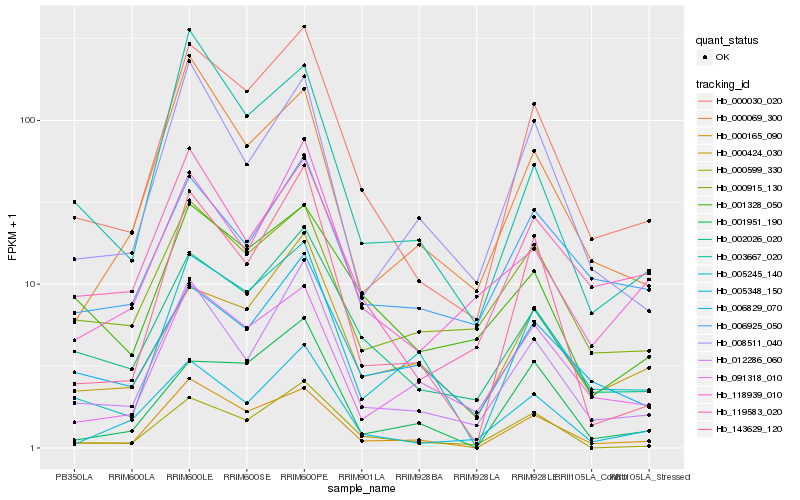
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 2 |
Hb_002026_020 |
0.1131513226 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 3 |
Hb_012286_060 |
0.1139494483 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000165_090 |
0.1150937644 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 5 |
Hb_005348_150 |
0.1304217792 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 6 |
Hb_000599_330 |
0.1360897777 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 7 |
Hb_005245_140 |
0.1371166383 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 8 |
Hb_143629_120 |
0.1394438451 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 9 |
Hb_119583_020 |
0.1517394107 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 10 |
Hb_000424_030 |
0.1535405061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_091318_010 |
0.1557864848 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 12 |
Hb_001951_190 |
0.1606792598 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 13 |
Hb_006829_070 |
0.1611985691 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 14 |
Hb_118939_010 |
0.1667132165 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_008511_040 |
0.1686206049 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_006925_050 |
0.1690812108 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 17 |
Hb_001328_050 |
0.1698009885 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 18 |
Hb_000915_130 |
0.1699474079 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 19 |
Hb_000069_300 |
0.1716076652 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_003667_020 |
0.1741448998 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |