| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
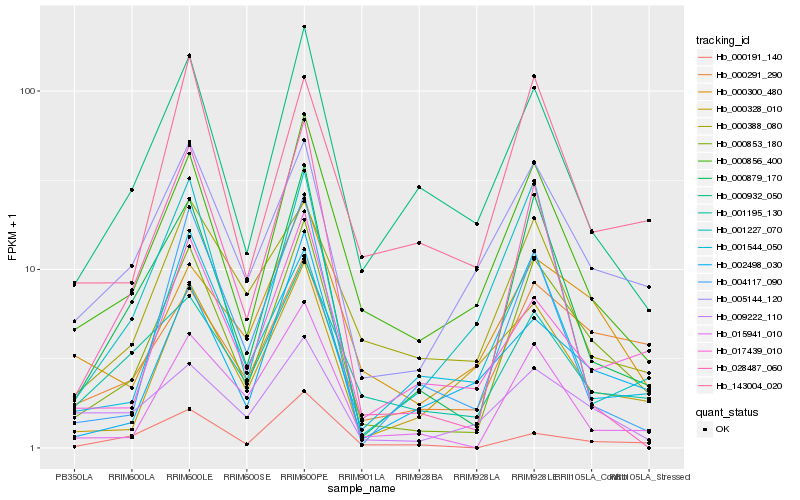
Hb_000853_180 |
0.0 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 2 |
Hb_000879_170 |
0.1265915053 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000856_400 |
0.1422963171 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 4 |
Hb_015941_010 |
0.1566418328 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_005144_120 |
0.1672335943 |
- |
- |
PREDICTED: probable glucuronoxylan glucuronosyltransferase IRX7 [Jatropha curcas] |
| 6 |
Hb_000291_290 |
0.1738736954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_017439_010 |
0.1777538376 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 8 |
Hb_000932_050 |
0.1803840309 |
- |
- |
annexin [Manihot esculenta] |
| 9 |
Hb_002498_030 |
0.1812078344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_143004_020 |
0.1812636194 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 11 |
Hb_000328_010 |
0.1831353397 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 12 |
Hb_001195_130 |
0.1837921046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004117_090 |
0.1889272483 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 14 |
Hb_000300_480 |
0.1911571639 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 15 |
Hb_009222_110 |
0.1926914171 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 16 |
Hb_001227_070 |
0.1940465488 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 17 |
Hb_028487_060 |
0.1945285626 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 18 |
Hb_001544_050 |
0.1950679074 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 19 |
Hb_000191_140 |
0.1966963026 |
- |
- |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000388_080 |
0.1967040318 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |