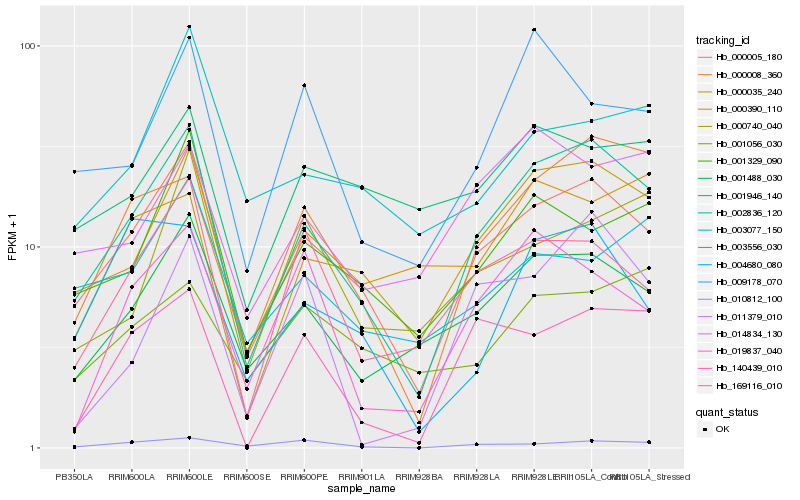
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002836_120 |
0.0 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 2 |
Hb_000005_180 |
0.0798530757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001488_030 |
0.1397942201 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 4 |
Hb_000008_360 |
0.1415560992 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 5 |
Hb_000390_110 |
0.1454069122 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 6 |
Hb_001329_090 |
0.1709790228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_169116_010 |
0.1748064104 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_140439_010 |
0.1768996653 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_014834_130 |
0.1815780073 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_009178_070 |
0.1837624712 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000035_240 |
0.184524949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010812_100 |
0.1895772323 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_019837_040 |
0.1958574294 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 14 |
Hb_004680_080 |
0.1979643384 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_000740_040 |
0.1983801934 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 16 |
Hb_003556_030 |
0.1988053724 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 17 |
Hb_011379_010 |
0.2023478493 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003077_150 |
0.2024040972 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 19 |
Hb_001946_140 |
0.2028086811 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001056_030 |
0.2067034203 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |