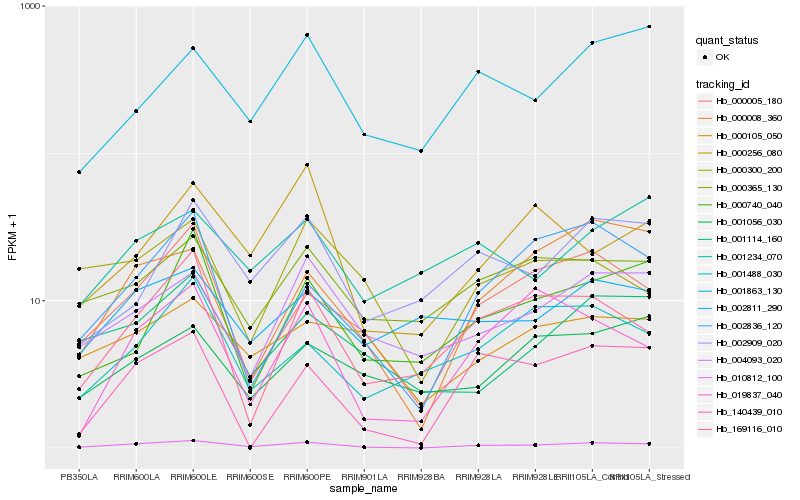
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_100 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_000005_180 |
0.1688341642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000008_360 |
0.1867910756 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 4 |
Hb_140439_010 |
0.1891147847 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_004093_020 |
0.1893691841 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 6 |
Hb_002836_120 |
0.1895772323 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 7 |
Hb_169116_010 |
0.1930015003 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_000365_130 |
0.1970695175 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 9 |
Hb_019837_040 |
0.1971588581 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 10 |
Hb_002909_020 |
0.2022977663 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 11 |
Hb_001863_130 |
0.2027492 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 12 |
Hb_001114_160 |
0.2086996725 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000105_050 |
0.2093971877 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 14 |
Hb_000300_200 |
0.2141661869 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000256_080 |
0.216062434 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 16 |
Hb_001488_030 |
0.2163646014 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 17 |
Hb_000740_040 |
0.2163884558 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 18 |
Hb_002811_290 |
0.2171066539 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 19 |
Hb_001056_030 |
0.2203777965 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001234_070 |
0.221995322 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |