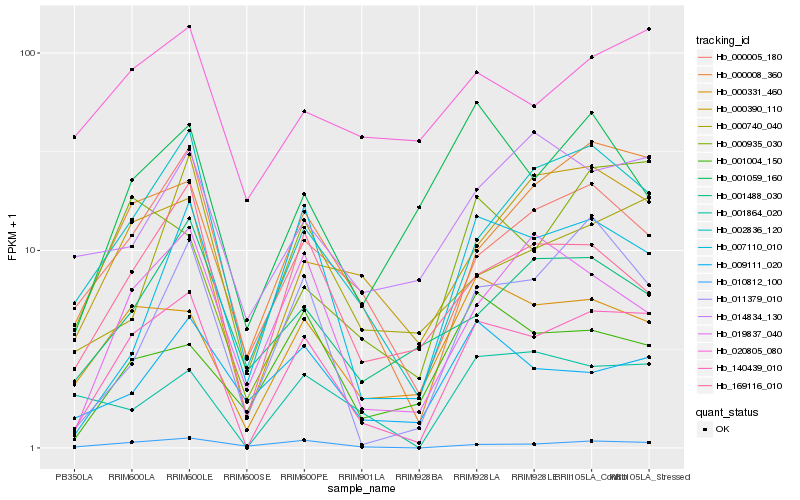
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140439_010 |
0.0 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_002836_120 |
0.1768996653 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 3 |
Hb_000008_360 |
0.1800122669 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 4 |
Hb_000331_460 |
0.1883159543 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 5 |
Hb_010812_100 |
0.1891147847 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 6 |
Hb_000005_180 |
0.196091146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_169116_010 |
0.1995118446 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_011379_010 |
0.2053568083 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000390_110 |
0.2062056667 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 10 |
Hb_007110_010 |
0.213813226 |
- |
- |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 11 |
Hb_000935_030 |
0.2146771087 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 12 |
Hb_019837_040 |
0.214846144 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 13 |
Hb_001864_020 |
0.2259099942 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000740_040 |
0.2298842821 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 15 |
Hb_001488_030 |
0.2322261806 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 16 |
Hb_009111_020 |
0.2336356178 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 17 |
Hb_001004_150 |
0.2359810511 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 18 |
Hb_001059_160 |
0.2367517745 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 19 |
Hb_020805_080 |
0.2381487776 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 20 |
Hb_014834_130 |
0.239456643 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |