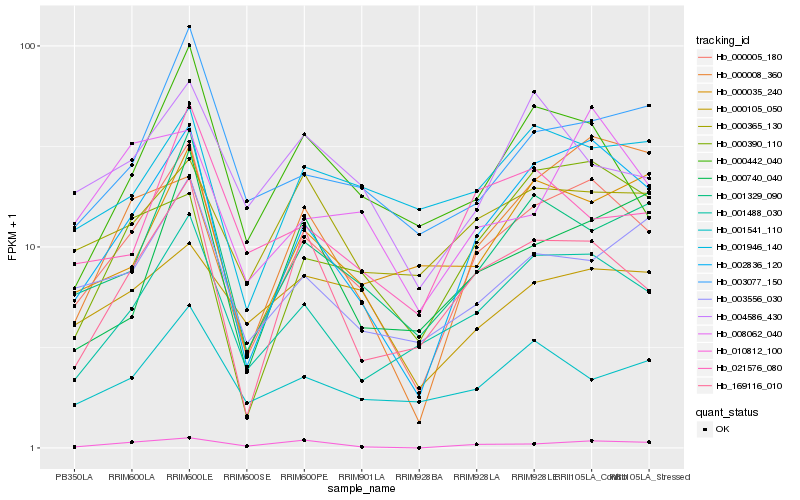
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002836_120 |
0.0798530757 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 3 |
Hb_001488_030 |
0.1295680639 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 4 |
Hb_001329_090 |
0.1403852695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_169116_010 |
0.1447628293 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_003077_150 |
0.1641713876 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 7 |
Hb_000442_040 |
0.1656837075 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003556_030 |
0.1681905917 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 9 |
Hb_010812_100 |
0.1688341642 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 10 |
Hb_000390_110 |
0.177164823 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 11 |
Hb_008062_040 |
0.1778366787 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001541_110 |
0.1792704979 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_001946_140 |
0.1793942995 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000035_240 |
0.1806439795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000365_130 |
0.1826953101 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 16 |
Hb_021576_080 |
0.1827798246 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 17 |
Hb_000008_360 |
0.1834350114 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 18 |
Hb_000740_040 |
0.1860829338 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 19 |
Hb_000105_050 |
0.1866373898 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 20 |
Hb_004586_430 |
0.1880327927 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |