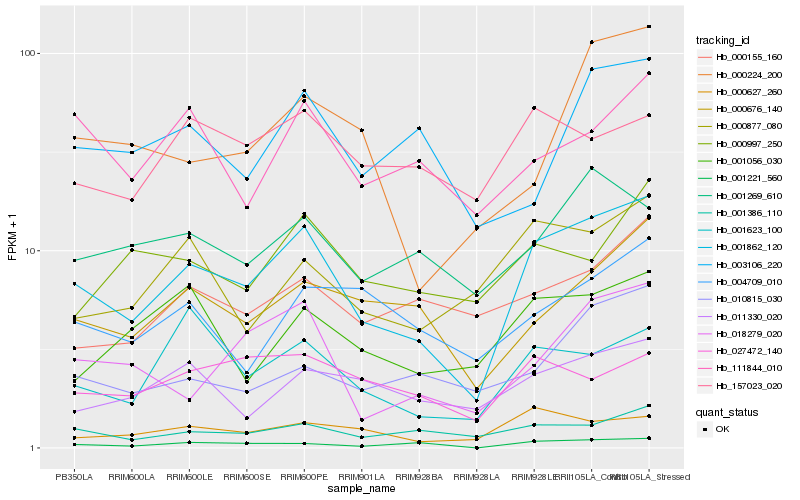
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_120 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 [Jatropha curcas] |
| 2 |
Hb_001386_110 |
0.1671556892 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 3 |
Hb_000676_140 |
0.1671822505 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 4 |
Hb_001221_560 |
0.1687068484 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase 1B-like [Jatropha curcas] |
| 5 |
Hb_000627_260 |
0.1754818042 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 6 |
Hb_001623_100 |
0.1771173455 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 7 |
Hb_018279_020 |
0.1784761618 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 8 |
Hb_000877_080 |
0.1798355302 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 9 |
Hb_011330_020 |
0.1834360578 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001269_610 |
0.1872074715 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_111844_010 |
0.1874327997 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 12 |
Hb_001056_030 |
0.1875961305 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004709_010 |
0.1903269972 |
- |
- |
hypothetical protein B456_007G097300 [Gossypium raimondii] |
| 14 |
Hb_010815_030 |
0.1911822278 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 15 |
Hb_003106_220 |
0.191236279 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_027472_140 |
0.1917310286 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646176 [Jatropha curcas] |
| 17 |
Hb_000155_160 |
0.1925047838 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 18 |
Hb_000224_200 |
0.1933769629 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 19 |
Hb_000997_250 |
0.1942218355 |
- |
- |
PREDICTED: probable alpha-mannosidase I MNS4 [Jatropha curcas] |
| 20 |
Hb_157023_020 |
0.1943666582 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |