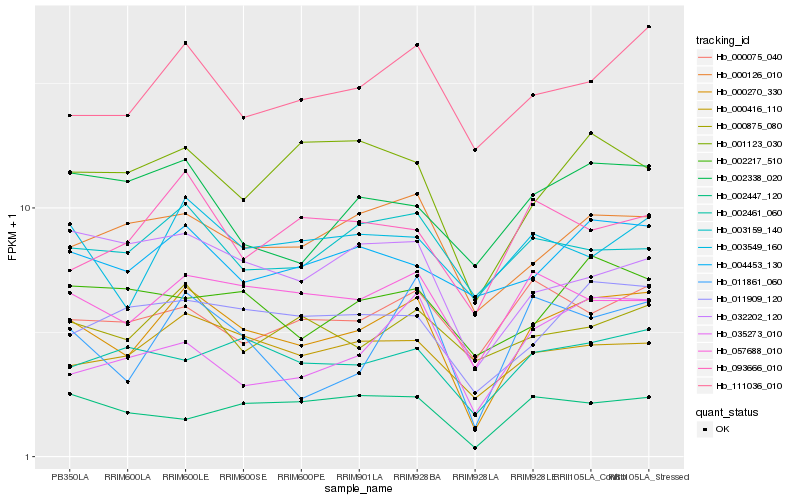
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_330 |
0.0 |
- |
- |
ribosomal protein S6 kinase, putative [Ricinus communis] |
| 2 |
Hb_111036_010 |
0.1228999209 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 3 |
Hb_000416_110 |
0.1239530497 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_057688_010 |
0.1263318734 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011909_120 |
0.1264491695 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000126_010 |
0.1288883326 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011861_060 |
0.1320897493 |
- |
- |
- |
| 8 |
Hb_002461_060 |
0.1331843768 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_003549_160 |
0.1351415863 |
- |
- |
Protein FAM18B, putative [Ricinus communis] |
| 10 |
Hb_001123_030 |
0.1395240193 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_032202_120 |
0.140977795 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004453_130 |
0.1417740622 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |
| 13 |
Hb_000875_080 |
0.1423081119 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_003159_140 |
0.1433725983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002217_510 |
0.1444495062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_093666_010 |
0.1448836708 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 17 |
Hb_002338_020 |
0.1452001654 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 18 |
Hb_000075_040 |
0.1453598039 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 19 |
Hb_035273_010 |
0.1454659603 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 20 |
Hb_002447_120 |
0.1459010216 |
- |
- |
hypothetical protein O988_01801, partial [Pseudogymnoascus pannorum VKM F-3808] |