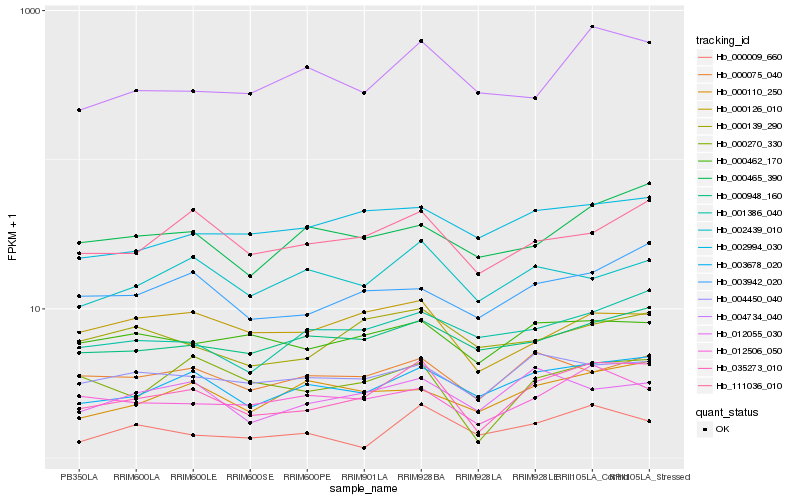
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035273_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 2 |
Hb_003678_020 |
0.121018787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_111036_010 |
0.1422336194 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 4 |
Hb_012506_050 |
0.1448602456 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000270_330 |
0.1454659603 |
- |
- |
ribosomal protein S6 kinase, putative [Ricinus communis] |
| 6 |
Hb_003942_020 |
0.1454889175 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004734_040 |
0.1471843956 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |
| 8 |
Hb_000075_040 |
0.1481891878 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 9 |
Hb_012055_030 |
0.1485171339 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 10 |
Hb_001386_040 |
0.1490911966 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 11 |
Hb_000948_160 |
0.1497191892 |
- |
- |
PREDICTED: 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004450_040 |
0.1507179535 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 13 |
Hb_000462_170 |
0.1522420883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000465_390 |
0.1535004611 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Jatropha curcas] |
| 15 |
Hb_000110_250 |
0.1537125477 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |
| 16 |
Hb_000139_290 |
0.1549238116 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002994_030 |
0.1567488866 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 18 |
Hb_000126_010 |
0.1574705538 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002439_010 |
0.1576194702 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 20 |
Hb_000009_660 |
0.1578907826 |
- |
- |
- |