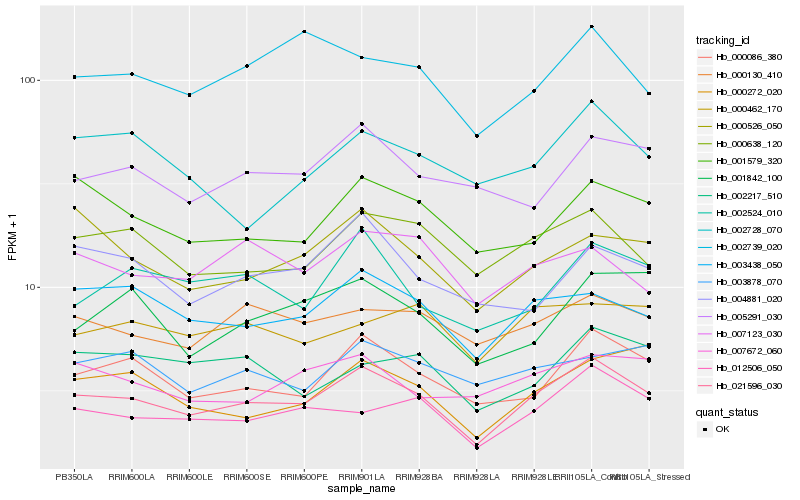
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021596_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000086_380 |
0.082560259 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 3 |
Hb_012506_050 |
0.0893875894 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003438_050 |
0.0913244434 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 5 |
Hb_000638_120 |
0.0914989869 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001579_320 |
0.1004148039 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 7 |
Hb_000130_410 |
0.100916967 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 8 |
Hb_002524_010 |
0.1041982106 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 9 |
Hb_005291_030 |
0.1053260029 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 10 |
Hb_000526_050 |
0.1060421816 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_007672_060 |
0.1068835989 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
Upstream activation factor subunit UAF30, putative [Ricinus communis] |
| 12 |
Hb_002739_020 |
0.1082665203 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 13 |
Hb_000272_020 |
0.1100535617 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 14 |
Hb_004881_020 |
0.1101322004 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 15 |
Hb_001842_100 |
0.1103550489 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 16 |
Hb_000462_170 |
0.1104520419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002217_510 |
0.1119829423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003878_070 |
0.1122282651 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 19 |
Hb_002728_070 |
0.112687694 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 20 |
Hb_007123_030 |
0.1130983967 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |