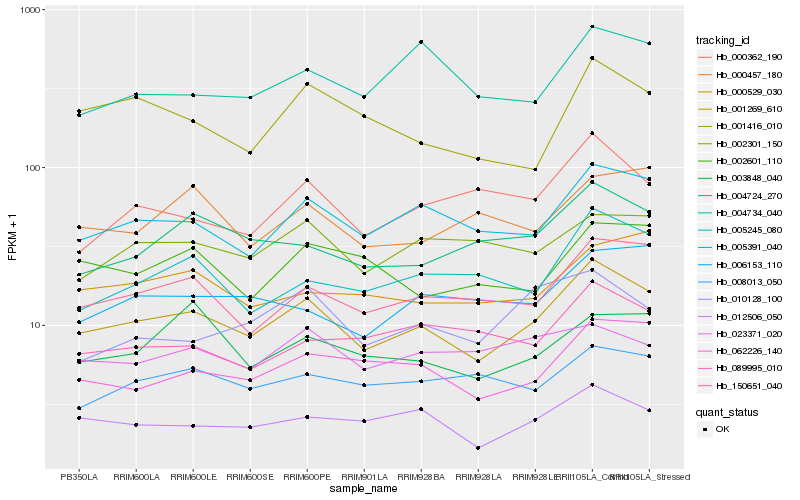
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_610 |
0.0 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_005391_040 |
0.0884103657 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA thioesterase 1 [Jatropha curcas] |
| 3 |
Hb_150651_040 |
0.0966417284 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 4 |
Hb_012506_050 |
0.1026848554 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_062226_140 |
0.1070735693 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |
| 6 |
Hb_004724_270 |
0.1146916878 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 7 |
Hb_005245_080 |
0.1157133447 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001416_010 |
0.1217297265 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 9 |
Hb_000362_190 |
0.1232911634 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 10 |
Hb_002301_150 |
0.1239901102 |
- |
- |
Drought-responsive family protein [Theobroma cacao] |
| 11 |
Hb_000529_030 |
0.1241012479 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_008013_050 |
0.1246700843 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 13 |
Hb_002601_110 |
0.1247187684 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 14 |
Hb_006153_110 |
0.1274200761 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_089995_010 |
0.1275229164 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like [Eucalyptus grandis] |
| 16 |
Hb_010128_100 |
0.127650015 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_023371_020 |
0.128778263 |
- |
- |
PREDICTED: uncharacterized protein LOC105633512 [Jatropha curcas] |
| 18 |
Hb_003848_040 |
0.1288756694 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_004734_040 |
0.1294853256 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |
| 20 |
Hb_000457_180 |
0.130925002 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |