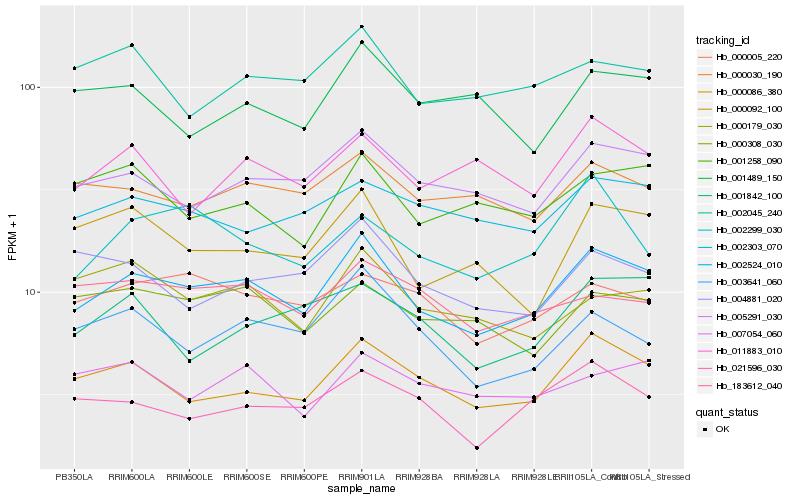
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002524_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s20250g [Populus trichocarpa] |
| 2 |
Hb_000086_380 |
0.0832591011 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 3 |
Hb_005291_030 |
0.0853349523 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 4 |
Hb_000092_100 |
0.101259916 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 5 |
Hb_000030_190 |
0.1020064652 |
- |
- |
PREDICTED: TATA-box-binding protein-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_003641_060 |
0.1022414379 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 7 |
Hb_021596_030 |
0.1041982106 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002303_070 |
0.1047155221 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 9 |
Hb_000005_220 |
0.1062884371 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 10 |
Hb_001258_090 |
0.1063226777 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 11 |
Hb_002045_240 |
0.1065832974 |
- |
- |
2-methyl-6-phytylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 12 |
Hb_000308_030 |
0.1079044725 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 13 |
Hb_011883_010 |
0.1106024572 |
- |
- |
PREDICTED: RNA-binding protein 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007054_060 |
0.1107275973 |
- |
- |
- |
| 15 |
Hb_004881_020 |
0.1107471956 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 16 |
Hb_001489_150 |
0.1108918807 |
- |
- |
PREDICTED: uncharacterized protein LOC105123623 isoform X2 [Populus euphratica] |
| 17 |
Hb_183612_040 |
0.1111745052 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 18 |
Hb_000179_030 |
0.1121072864 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002299_030 |
0.1133115601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001842_100 |
0.1141585815 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |