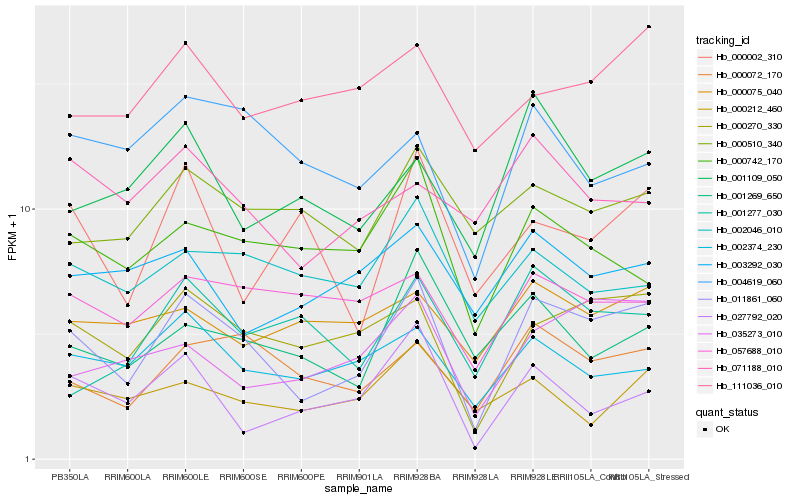
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_000270_330 |
0.1320897493 |
- |
- |
ribosomal protein S6 kinase, putative [Ricinus communis] |
| 3 |
Hb_000072_170 |
0.1530398983 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 4 |
Hb_035273_010 |
0.1668730803 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 5 |
Hb_002374_230 |
0.1711618998 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 6 |
Hb_001269_650 |
0.1790452409 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_000742_170 |
0.1803210691 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_001109_050 |
0.180790735 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |
| 9 |
Hb_057688_010 |
0.1815249525 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000510_340 |
0.183393674 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 11 |
Hb_027792_020 |
0.1838232916 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 12 |
Hb_111036_010 |
0.1850544343 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 13 |
Hb_002046_010 |
0.1866773993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000075_040 |
0.1873158733 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 15 |
Hb_003292_030 |
0.188000091 |
- |
- |
PREDICTED: exosome complex component RRP42 [Populus euphratica] |
| 16 |
Hb_001277_030 |
0.1896143833 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 17 |
Hb_000212_460 |
0.1907574904 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 18 |
Hb_004619_060 |
0.1908976045 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 19 |
Hb_071188_010 |
0.1909896463 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 20 |
Hb_000002_310 |
0.1914881917 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |