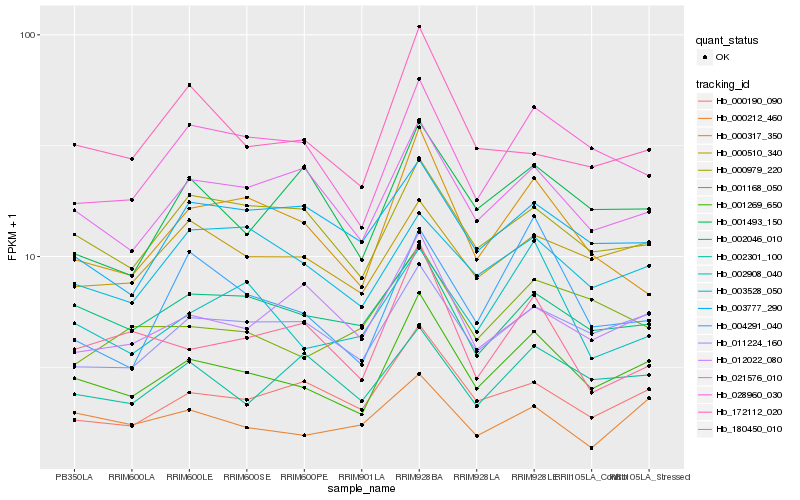
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_650 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_011224_160 |
0.0975761465 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 3 |
Hb_021576_010 |
0.1040783175 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 4 |
Hb_000190_090 |
0.1070949674 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 5 |
Hb_028960_030 |
0.1080490932 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 6 |
Hb_000979_220 |
0.1128198676 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000510_340 |
0.1133339899 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 8 |
Hb_002046_010 |
0.1137479539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000317_350 |
0.1185736483 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 10 |
Hb_000212_460 |
0.1192883201 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 11 |
Hb_002908_040 |
0.1237541827 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_002301_100 |
0.1252232508 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 13 |
Hb_001493_150 |
0.1255156371 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 14 |
Hb_012022_080 |
0.1258756415 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_003528_050 |
0.126282556 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_180450_010 |
0.1278186456 |
- |
- |
FF domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_172112_020 |
0.1281401927 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 18 |
Hb_001168_050 |
0.1287212038 |
- |
- |
hypothetical protein POPTR_0007s03190g [Populus trichocarpa] |
| 19 |
Hb_004291_040 |
0.1294110953 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_003777_290 |
0.1294409673 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |