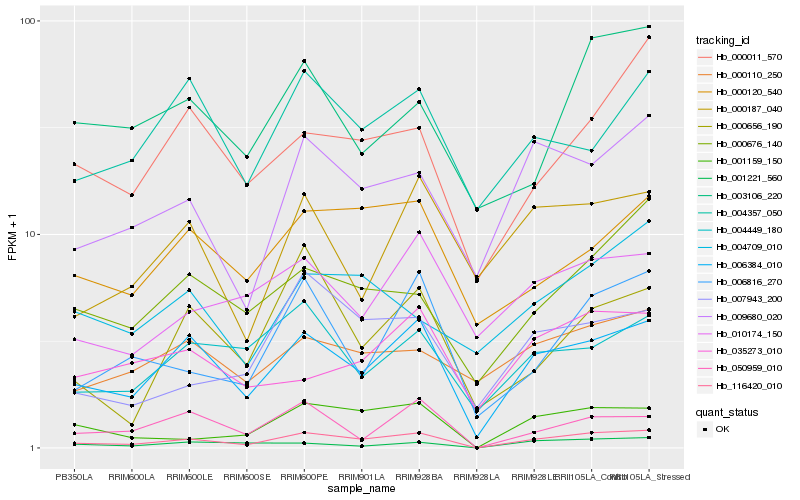
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116420_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_006384_010 |
0.1039100605 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_001159_150 |
0.1494662934 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_006816_270 |
0.1710828566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009680_020 |
0.1713938676 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 6 |
Hb_007943_200 |
0.1828003287 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_000676_140 |
0.1837459032 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 8 |
Hb_050959_010 |
0.1840878984 |
- |
- |
- |
| 9 |
Hb_004449_180 |
0.1867541246 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 10 |
Hb_000656_190 |
0.1898689304 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |
| 11 |
Hb_000011_570 |
0.1907900902 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 12 |
Hb_000120_540 |
0.1909232499 |
- |
- |
translation initiation factor, putative [Ricinus communis] |
| 13 |
Hb_001221_560 |
0.1932904024 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase 1B-like [Jatropha curcas] |
| 14 |
Hb_010174_150 |
0.1960537051 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_003106_220 |
0.1983434501 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_035273_010 |
0.1996507815 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 17 |
Hb_000187_040 |
0.1997976254 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 18 |
Hb_004357_050 |
0.2000480636 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 19 |
Hb_000110_250 |
0.2001021099 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |
| 20 |
Hb_004709_010 |
0.2041810833 |
- |
- |
hypothetical protein B456_007G097300 [Gossypium raimondii] |