| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
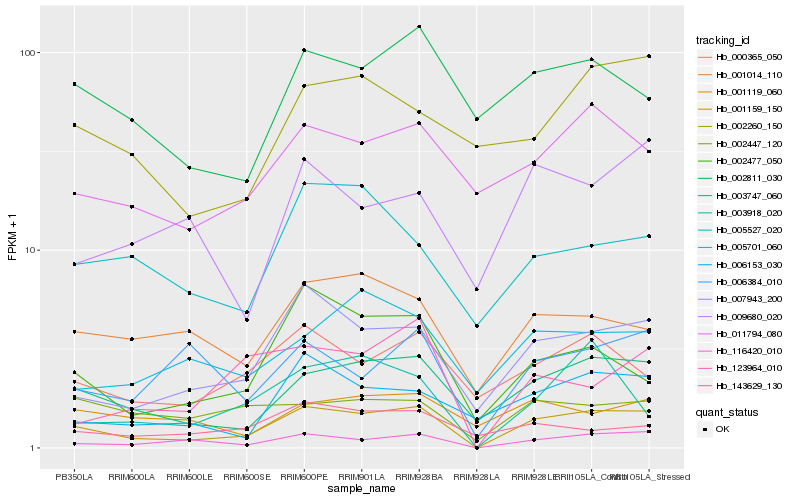
Hb_001159_150 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_116420_010 |
0.1494662934 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 3 |
Hb_003747_060 |
0.1495489283 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 4 |
Hb_007943_200 |
0.1608567687 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_002477_050 |
0.1935583011 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000365_050 |
0.1978454123 |
- |
- |
- |
| 7 |
Hb_011794_080 |
0.2003146315 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 8 |
Hb_006153_030 |
0.201359531 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 9 |
Hb_001014_110 |
0.2021237595 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 10 |
Hb_006384_010 |
0.2035452714 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_123964_010 |
0.2091389648 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 12 |
Hb_002811_030 |
0.2095115728 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 13 |
Hb_002447_120 |
0.2126559401 |
- |
- |
hypothetical protein O988_01801, partial [Pseudogymnoascus pannorum VKM F-3808] |
| 14 |
Hb_005701_060 |
0.2129497264 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 15 |
Hb_005527_020 |
0.2142497332 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 16 |
Hb_003918_020 |
0.2165525117 |
- |
- |
- |
| 17 |
Hb_002260_150 |
0.2166371772 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 18 |
Hb_143629_130 |
0.216984726 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_001119_060 |
0.2175415555 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 20 |
Hb_009680_020 |
0.2177646666 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |