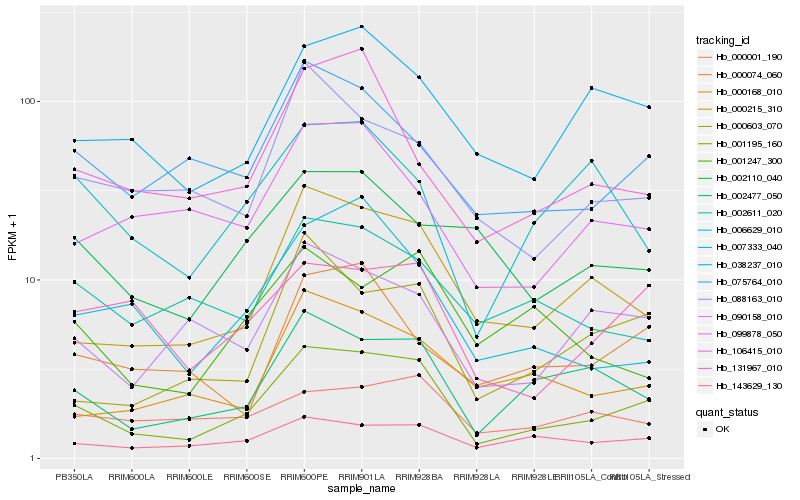
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_002477_050 |
0.1505773187 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_000215_310 |
0.1507699588 |
- |
- |
- |
| 4 |
Hb_038237_010 |
0.1681763425 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 5 |
Hb_143629_130 |
0.1724795181 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_090158_010 |
0.1737656521 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 7 |
Hb_131967_010 |
0.1765104062 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 8 |
Hb_000603_070 |
0.1777589522 |
- |
- |
- |
| 9 |
Hb_075764_010 |
0.1785209376 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 10 |
Hb_007333_040 |
0.1845884013 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 11 |
Hb_000168_010 |
0.188265184 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 12 |
Hb_088163_010 |
0.188448834 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 13 |
Hb_099878_050 |
0.1924774836 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 14 |
Hb_002611_020 |
0.1934330079 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 15 |
Hb_106415_010 |
0.1942070684 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 16 |
Hb_006629_010 |
0.1972719474 |
- |
- |
Glutathione S-transferase 2 isoform 3 [Theobroma cacao] |
| 17 |
Hb_000074_060 |
0.1996493989 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 18 |
Hb_000001_190 |
0.1996939645 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 19 |
Hb_002110_040 |
0.1997236105 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 20 |
Hb_001247_300 |
0.1999924425 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |