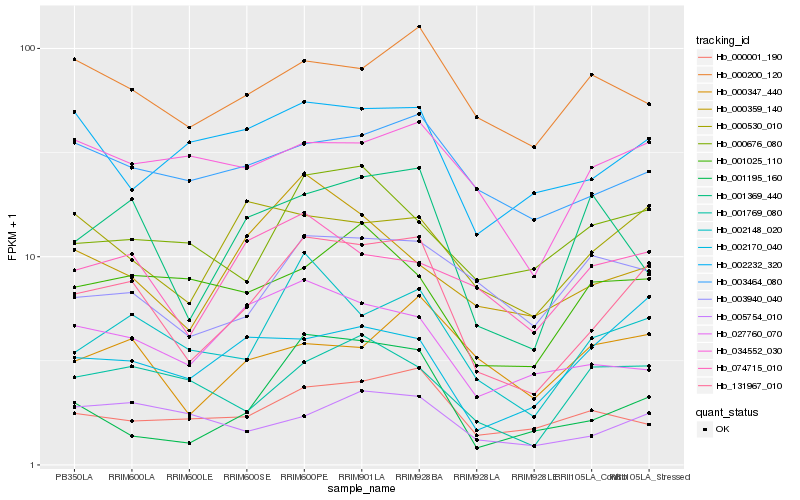
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131967_010 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_002148_020 |
0.130794616 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 3 |
Hb_000001_190 |
0.1679671323 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 4 |
Hb_005754_010 |
0.1699065988 |
- |
- |
hypothetical protein POPTR_0660s00210g [Populus trichocarpa] |
| 5 |
Hb_003464_080 |
0.1707778137 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 6 |
Hb_000530_010 |
0.1709122937 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 7 |
Hb_001769_080 |
0.17301821 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |
| 8 |
Hb_001369_440 |
0.1730442584 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 9 |
Hb_074715_010 |
0.1746523824 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002170_040 |
0.1753489736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001195_160 |
0.1765104062 |
- |
- |
- |
| 12 |
Hb_000676_080 |
0.1770191344 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 13 |
Hb_000347_440 |
0.1776339091 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 14 |
Hb_000359_140 |
0.177744507 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 15 |
Hb_034552_030 |
0.1780118472 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_003940_040 |
0.1781312625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001025_110 |
0.1785251617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_027760_070 |
0.1787066747 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 19 |
Hb_000200_120 |
0.1790808014 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002232_320 |
0.1793671269 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |